2012 Annual Science Report
 University of Hawaii, Manoa
Reporting | SEP 2011 – AUG 2012
University of Hawaii, Manoa
Reporting | SEP 2011 – AUG 2012
Charting the Universe of Amino Acid Structures
Project Summary
More than 3.5 billion years ago, life on our planet evolved a precise alphabet of 20 amino acids to function as building blocks which cells use to construct proteins according to genetic instructions. However, the twenty genetically encoded amino acids are but a tiny fraction of the chemical structures that could plausibly play such a role. Any science of the origins, distribution and future of life in the universe must take into account this larger context of chemical structures. But while astrochemistry, prebiotic chemistry, and bioengineering all hint at the chemical structures it contains, until now this amino acid universe has remained largely unexplored. Efforts to describe the structures it contains, or even estimate their number, have been hampered by the complexity inherent to the combinatorial properties of organic molecules. We have formed a new collaboration to combine European (DLR) advances in computational chemistry with NAI expertise in organic chemistry and amino acid biology to address this gap in current scientific understanding. Our early results have provided the first ever sketch of the amino acid structure universe, showing it to be far larger and more complex than previously supposed. This forms an important milestone in defining and exploring the principles of “universal biology”
Project Progress
In this project we formed a new, interdisciplinary collaboration that used sophisticated computer software based on graph theory and constructive combinatorics in order to conduct an efficient and exhaustive search of the chemical structures implied by three different approaches to defining the set of α-amino acids relevant to coded biological proteins. Our results include virtual libraries of α-amino acid chemical structures corresponding to these different approaches, comprising 121,614 and 3,946 structures respectively. Most importantly, this work points to the the existence of much larger, as yet uncomputed libraries, and suggests how these refined explorations might best proceed. We are currently writing up these results as a first ever exploration of the true complexity of the amino acid structure universe. Our report describes the methods and computational resources by which these libraries were generated, and the chemical structures obtained and discusses how these different approaches can be used with respect to future applications, including future efforts to explore the remaining, uncharted regions of the amino acid structure universe.
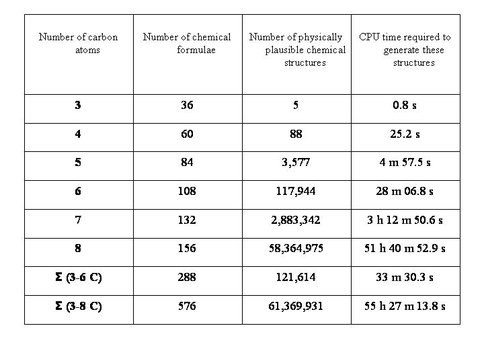
This table shows the number of unique chemical structures for a simple definition of “biologically plausible alpha amino acid” as a function of the size of the side-chain (measured as number of C atoms). Total structures refers to all structures that satisfy the fundamental rules of chemistry (covalent bonding); plausible structures refers to a refined search that excludes classes of sub-structure known to be unstable or otherwise unfavorable for reasons of physics and chemistry. Processing times refer to MOLGEN 5.01 running under Linux on an Intel Dual Core CPU running at 2.66 GHz.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Stephen Freeland
Project Investigator
Henderson Cleaves
Co-Investigator
Markus Meringer
Collaborator
-
RELATED OBJECTIVES:
Objective 3.1
Sources of prebiotic materials and catalysts
Objective 3.2
Origins and evolution of functional biomolecules
Objective 6.2
Adaptation and evolution of life beyond Earth
Objective 7.1
Biosignatures to be sought in Solar System materials
Objective 7.2
Biosignatures to be sought in nearby planetary systems