2012 Annual Science Report
 Massachusetts Institute of Technology
Reporting | SEP 2011 – AUG 2012
Massachusetts Institute of Technology
Reporting | SEP 2011 – AUG 2012
The Development of Sensory and Nervous Systems in the Basal Branches of the Animal Tree
Project Summary
Animals interact with the world through complex sensory structures (eyes, ears, antennas, etc.), which are coordinated by collections of neurons. While the nervous and sensory systems of animals are incredibly diverse, a growing body of evidence suggests that many of these systems are controlled by similar sets of genes. We are looking at early branching and understudied lineages of the animal family tree (using the jellyfish Aurelia and the worm Neanthes respectively) to see if these animals use similar genes during neurosensory development as the better-studied fruit fly and mouse. This research is critical for determining which structures are shared between animals because of common ancestry (known as homologous structures) and those that evolved independently in different lineages. Ultimately, such research informs how morphologically and behaviorally complex animals evolve.
Project Progress
The range of work under development in the lab is quite exciting. First off, we are sequencing the genome of the jellyfish Aurelia. This work involves collaboration between researchers at UC San Diego, UC Irvine, and UC Santa Barbara as well as at UCLA. We now have on the order of 100X coverage of the genome with Illumina paired-end reads, and Pac Bio sequencing to aid in the assembly of larger contigs is proceeding. This genome provides a significant advance in our ability to unravel the early evolution of complex life, as it permits us to compare the cnidarians in far greater detail, significantly clarifying the evolutionary complexity at the inception of the bilaterian clade, and permitting detailed work comparing Cnidaria with and without the sense organ rich “medusa” (or jellyfish) phase. This will be a tremendous boon to our work on basal metazoan sense organs.
In our lab we’ve extensively sequenced RNA (cDNA), and we now have a proteome in hand. Using this backbone, we are proceeding with efforts to conduct assessments of differential gene expression through life history stages and the formation of sense organs, using RNA-Seq alongside gene expression data (Figure 1). Finally, we have engaged in collaborative research on the microRNA component of Aurelia with the Peterson lab. These genomic approaches, in combination and in collaboration with our NAI colleagues, are sure to bear exciting results relative to the understanding of the evolution of eukaryotic multicellular complexity generally, and sense organ development specifically.
We have papers in press on the expression of two LIM homeobox genes in a polychaete annelid. The genes examined regulate the differentiation of neural/sensory /appendage structures in many animals, but have not been explored in representatives of basal Lophotrochozoa. This work helps clarify the early evolution of Bilaterian neural sensory complexity. In addition, we have work in press examining the evolution of stem cells in basal Metazoa, in particular the Cnidaria. Stem cells are a topic of considerable interest from a biomedical standpoint. Here we examine their evolution at the base of the Metazoa, an exercise critical to placing the biomedical exercise in evolutionary context as well as understanding the evolution of cellular complexity across animals.
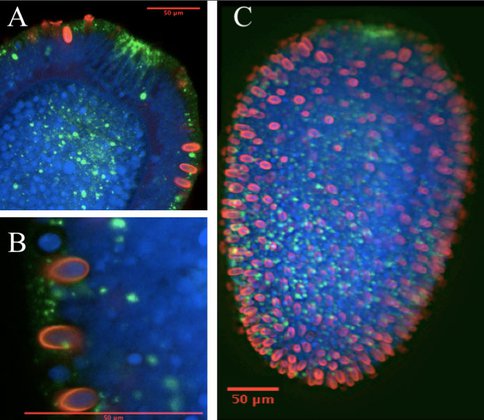
An example of gene expression data collected this year. A planula larvae of Aurelia, with PAX expression in green, stinging cells (cnidocytes) labeled in red, and cell nuclei labeled in blue. Close-ups in (A) and (B).
Publications
-
Gold, D. A., & Jacobs, D. K. (2012). Stem cell dynamics in Cnidaria: are there unifying principles?. Dev Genes Evol, 223(1-2), 53–66. doi:10.1007/s00427-012-0429-1
-
Takashima, S., Gold, D., & Hartenstein, V. (2012). Stem cells and lineages of the intestine: a developmental and evolutionary perspective. Dev Genes Evol, 223(1-2), 85–102. doi:10.1007/s00427-012-0422-8
-
Winchell, C. J., & Jacobs, D. K. (2013). Expression of the Lhx genes apterous and lim1 in an errant polychaete: implications for bilaterian appendage evolution, neural development, and muscle diversification. EvoDevo, 4(1), 4. doi:10.1186/2041-9139-4-4
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Nagayasu Nakanishi
Co-Investigator
Chris Winchell
Co-Investigator
Kevin Peterson
Collaborator
Kira Cozzolino
Undergraduate Student
José López
Undergraduate Student
-
RELATED OBJECTIVES:
Objective 4.1
Earth's early biosphere.
Objective 4.2
Production of complex life.