2012 Annual Science Report
 Massachusetts Institute of Technology
Reporting | SEP 2011 – AUG 2012
Massachusetts Institute of Technology
Reporting | SEP 2011 – AUG 2012
Reconstruction of Ancient Proteins
Project Summary
The genetic code is one of the most ancient and universal aspects of biology on Earth, and determines how specific DNA sequences get interpreted as peptide sequences, which then fold into all the proteins necessary for the growth and function of living cells. To a large extent, this code is determined by a class of proteins that specify which RNA adaptor molecules (tRNA) become attached to which amino acids, aminoacyl-tRNA synthetases. Therefore, reconstructing the amino acid sequences of the ancestors of these synthetases, existing ~4 billion years ago, can tell us the mechanisms by which the genetic code arose, and how it evolved to the modern form inherited by all known living organisms.
Project Progress
Our primary research project has been the ancestral reconstruction of ancient protein sequences early in the history of life, in order to elucidate primordial events in the development of the genetic code. We have developed novel methods in ancestral reconstruction using detailed biological information, specifically, horizontal gene transfer, intragenic recombination, complex models of sequence evolution, and protein structure information. One of the most important parts of protein synthesis is the aminoacylation of tRNA with the correct amino acid, a step that defines the syntax of the genetic code, mediated by a related set of ancient protein families, aminoacyl-tRNA synthetases (aaRS). In this approach, we assume that the sequence of the reconstructed ancestors of aaRS proteins should show an absence of usage of their cognate amino acids, if the mechanism of the addition of these amino acids was the divergence of the synthetases in question. Conversely, if the cognate amino acids of groups of synthetases are inferred to be present within ancestor sequences before their divergences, the use of these amino acids within proteins must predate their protein-mediated incorporation, directly implying a more primitive system for enforcing the genetic code at earlier stages in protein evolution.
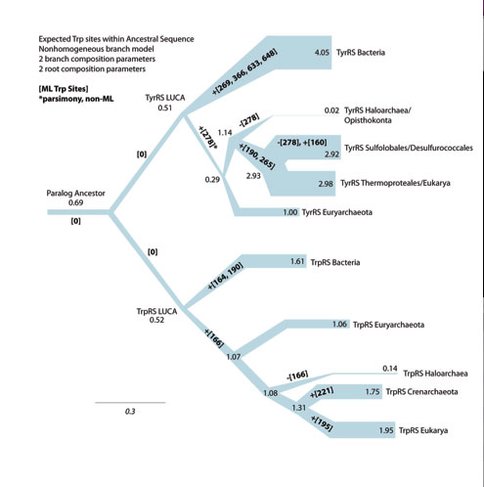
We have identified strong evidence that some parts of the genetic code, such as the usage of the hydrophobic amino acids isoleucine and valine, predate the protein machinery for their incorporation, and were likely invented during an early time when an RNA-based physiology was still preeminent. Conversely, tryptophan seems to be a more recent addition, with a conspicuous absence in the deep protein ancestors of the enzymes responsible for incorporating Trp in the code (Figure 1). These results are currently being verified via in silico simulations, and developing in vitro methodologies for testing the functionality of synthesized ancestral protein variants, with collaborators at Harvard University.
In the course of this work, several discoveries about the extent of horizontal gene transfer(HGT)-associated recombination were made, which are actively being investigated as to their impact on the inferred topology of the Tree of Life. Reconciling HGT events in this manner has also led to two additional projects: the characterization of a novel anti-protozoan drug target evolving via ancient HGT, the first of its kind to emerge from paleogenomic/astrobiological research; and a novel microbial ecology explanation for the End-Permian mass extinction, via the emergence of a globally dominant methanogenic pathway, evolving via HGT. The investigation of this novel drug target is still in an early bioinformatics-based stage, while the research into the microbial cause of the mass extinction is complete in its current phase, and in the final stages of manuscript preparation for publication.
Ancestral reconstructions of the two related aminoacyl-tRNA protein families responsible for the use of tryptophan and tyrosine in the genetic code, with respect to tryptophan content across the inferred ancestors within each node of the Tree of Life for each protein. “LUCA” nodes denote the “Last Universal Common Ancestor” of life, which contained a version of each synthetase paralog. “Paralog Ancestor” is the common ancestor of TrpRS and TyrRS protein families, which diverged before LUCA. Branch thickness relates to the expected Trp content (number of sites) of the ancestor protein sequence at each node (as numbered). Bold numbers within branches relate to the specific sites within the protein which were inferred to gain (+) or lose (-) a Trp residue along each branch of the Tree of Life within each gene family. The reconstruction shows that all sites containing Trp within these proteins arose at a relatively late time, after LUCA, and independently across many groups of organisms, supporting the absence of Trp within the paralog ancestor and its immediate descendants (as indicated by [0] sites).
Publications
-
Fournier, G. P., Andam, C. P., Alm, E. J., & Peter Gogarten, J. (2011). Molecular Evolution of Aminoacyl tRNA Synthetase Proteins in the Early History of Life. Orig Life Evol Biosph, 41(6), 621–632. doi:10.1007/s11084-011-9261-2
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Eric Alm
Co-Investigator
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules
Objective 3.4
Origins of cellularity and protobiological systems
Objective 4.1
Earth's early biosphere.
Objective 4.2
Production of complex life.