2012 Annual Science Report
 Massachusetts Institute of Technology
Reporting | SEP 2011 – AUG 2012
Massachusetts Institute of Technology
Reporting | SEP 2011 – AUG 2012
Molecular Basis for Complexity Development
Project Summary
Cadherins are large, multi-domain proteins that are also cell surface receptors which function in cell adhesion, cell polarity, and tissue morphogenesis. They are considered essential to the appearance of animal life. We found cadherin genes in Capsaspora owczarzaki and the choanoflagellate Salpingoeca rosetta suggesting that this protein family predates the divergence of the C. owczarzaki, choanoflagellate, and metazoan lineages.
Project Progress
Cadherins are large, multi-domain proteins that are also cell surface receptors which function in cell adhesion, cell polarity, and tissue morphogenesis. They are considered essential to the appearance of animal life. We found cadherin genes in Capsaspora owczarzaki and the choanoflagellate Salpingoeca rosetta suggesting that this protein family predates the divergence of the C. owczarzaki, choanoflagellate, and metazoan lineages.
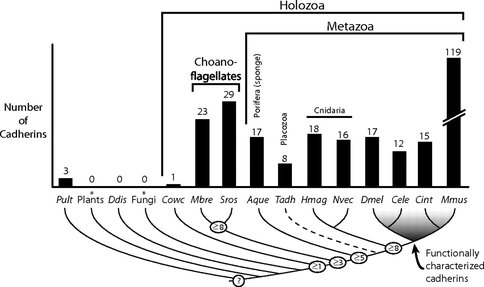
The evolution of cadherins, which are essential for metazoan multicellularity and restricted to metazoans and their closest relatives, has special relevance for understanding metazoan origins. To reconstruct the ancestry and evolution of cadherin gene families, we analyzed the genomes of the choanoflagellate Salpingoeca rosetta, the unicellular outgroup of choanoflagellates and metazoans Capsaspora owczarzaki, and a draft genome assembly from the homoscleromorph sponge Oscarella carmela. Our finding of a cadherin gene in C. owczarzaki reveals that cadherins predate the divergence of the C. owczarzaki, choanoflagellate, and metazoan lineages. Data from these analyses also suggest that the last common ancestor of metazoans and choanoflagellates contained representatives of at least three cadherin families, lefftyrin, coherin, and hedgling. Additionally, we find that an O. carmela classical cadherin has predicted structural features that, in bilaterian classical cadherins, facilitate binding to the cytoplasmic protein β-catenin and, thereby, promote cadherin-mediated cell adhesion. In contrast with premetazoan cadherin families (i.e., those conserved between choanoflagellates and metazoans), the later appearance of classical cadherins coincides with metazoan origins.
Phylogenetic distribution and abundance of cadherins in the genomes of diverse eukaryotes. Once thought to be restricted to metazoans, cadherins are abundant in choanoflagellates and evolved before the divergence of Capsaspora owczarzaki, choanoflagellates, and metazoans . EC domains detected in the genome of the oomycte Pythium ultimum likely evolved through convergence or lateral gene transfer (9). The number of cadherin families inferred at ancestral nodes (determined based upon their shared domain composition and organization) is indicated (open circles). The dashed lineage of Trichoplax adhaerens reflects its uncertain phylogenetic placement. Aque, A. queenslandica; Cele, Caenorhabditis elegans; Cint, Ciona intestinalis; Cowc, C. owczarzaki; Ddis, Dictyostelium discoideum; Dmel, D. melanogaster; Hmag, Hydra magnipapillata; Mbre, M. brevicollis; Mmus, Mus musculus; Nvec, N. vectensis; Pult, P. ultimum; Sros, S. rosetta; Tadh, T. adhaeren
Publications
-
Alegado, R. A., Grabenstatter, J. D., Zuzow, R., Morris, A., Huang, S. Y., Summons, R. E., & King, N. (2012). Algoriphagus machipongonensis sp. nov., co-isolated with a colonial choanoflagellate. INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY, 63(Pt 1), 163–168. doi:10.1099/ijs.0.038646-0
-
Nichols, S. A., Roberts, B. W., Richter, D. J., Fairclough, S. R., & King, N. (2012). Origin of metazoan cadherin diversity and the antiquity of the classical cadherin/ -catenin complex. Proceedings of the National Academy of Sciences, 109(32), 13046–13051. doi:10.1073/pnas.1120685109
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Rosanna Alegardo
Postdoc
-
RELATED OBJECTIVES:
Objective 4.2
Production of complex life.