2012 Annual Science Report
 Massachusetts Institute of Technology
Reporting | SEP 2011 – AUG 2012
Massachusetts Institute of Technology
Reporting | SEP 2011 – AUG 2012
Micro-RNAs as Phylogenetic Markers
Project Summary
Since the beginning of 2011, we have published 12 peer-reviewed papers centered on three themes. First, we have pioneered the use of microRNAs (miRNAs) – small 22 nucleotide non-coding RNAs – as phylogenetic characters, and have recently gained new insights into the relationships of vertebrates, arthropods, brachiopods, and flies. Second, we have noted that the number of miRNAs an organism possesses seems related to their relative morphological complexity such that more complex animals have many more miRNAs than simple animals. Third, in a recent Science paper (Erwin et al.2012) we summarized all of our molecular-clock work to date which strongly suggests that bilaterians have a fairly extensive cryptic Precambrian history, which when coupled with our miRNA work makes this missing record even more enigmatic, and the resolution of this paradox is the focus of our current and much of our future NASA-sponsored work.
Project Progress
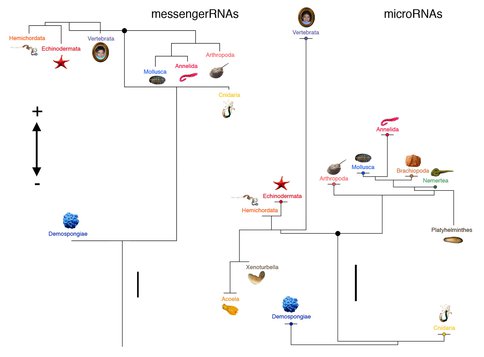
During the last year we have published 7 papers using microRNAs to help elucidate phylogenetic relationships that have thus far proved difficult using standard molecular means. Questions addressed range from the position of turtles within the reptiles (Lyson et al. 2011) to the monophyly of brachiopods relative to phoronids (Sperling et al. 2011) to the phylogenetic position of tardigrades relative to other ecdysozoans (Campbell et al. 2011). We have further used miRNAs to help test hypotheses concerning the advent and maintenance of morphological complexity. We have shown that complex animals have many more miRNAs than simple animals (see Fig. 1), and that secondary morphological simplification is accompanied by secondary loss of miRNAs. In particular we showed that acoel flatworms, long a proxy for a “typical” Precambrian bilaterian animal as they were reconstructed as basal bilaterians with a relatively simple morphology, were in fact closely related to modern hemichordates and echinoderms (Philippe et al. 2011), and thus their simple morphology was secondarily derived through loss, including the loss of a centralized nervous system, blood vascular system, and kidney. Further, this secondary simplification was accompanied by a dramatic loss of miRNAs (see Fig. 1). Current work is focused on further testing this correlation between morphological simplicity/complexity and miRNA repertoire.
One hundred and thirty-one representative transcription factors and signaling ligands were coded for eight metazoan taxa and mapped onto a widely accepted metazoan topology (left). The length of the branch represents the total amount of mRNA genes acquired minus those that were lost (scale bar represents ten genes total). Much of the developmental mRNA toolkit was acquired before the last common ancestor of cnidarians and bilaterians. This is in contrast to the miRNA repertoire that displays extensive gain of miRNAs in the bilaterian stem lineage after it split from cnidarians (right). All 139 miRNA families known from 22 metazoan species were coded, and similar to the mRNA figure (left) the length of the branch represents the total amount of miRNA genes gained at that point minus those that were secondarily lost (scale bar = ten genes total). Note that increases to morphological complexity are correlated with increases to the miRNA toolkit, and secondary simplifications in morphology correlate with a relatively high level of secondary miRNA loss. From Erwin et al. (2011).
Publications
-
Anderson, B. M., Pisani, D., Miller, A. I., & Peterson, K. J. (2011). The environmental affinities of marine higher taxa and possible biases in their first appearances in the fossil record. Geology, 39(10), 971–974. doi:10.1130/g32413.1
-
Campbell, L. I., Rota-Stabelli, O., Edgecombe, G. D., Marchioro, T., Longhorn, S. J., Telford, M. J., … Pisani, D. (2011). MicroRNAs and phylogenomics resolve the relationships of Tardigrada and suggest that velvet worms are the sister group of Arthropoda. Proceedings of the National Academy of Sciences, 108(38), 15920–15924. doi:10.1073/pnas.1105499108
-
Campo-Paysaa, F., Sémon, M., Cameron, R. A., Peterson, K. J., & Schubert, M. (2011). microRNA complements in deuterostomes: origin and evolution of microRNAs. Evolution & Development, 13(1), 15–27. doi:10.1111/j.1525-142×.2010.00452.x
-
Erwin, D. H., Laflamme, M., Tweedt, S. M., Sperling, E. A., Pisani, D., & Peterson, K. J. (2011). The Cambrian Conundrum: Early Divergence and Later Ecological Success in the Early History of Animals. Science, 334(6059), 1091–1097. doi:10.1126/science.1206375
-
Lyson, T. R., Sperling, E. A., Heimberg, A. M., Gauthier, J. A., King, B. L., & Peterson, K. J. (2011). MicroRNAs support a turtle + lizard clade. Biology Letters, 8(1), 104–107. doi:10.1098/rsbl.2011.0477
-
Philippe, H., Brinkmann, H., Copley, R. R., Moroz, L. L., Nakano, H., Poustka, A. J., … Telford, M. J. (2011). Acoelomorph flatworms are deuterostomes related to Xenoturbella. Nature, 470(7333), 255–258. doi:10.1038/nature09676
-
Pisani, D., Feuda, R., Peterson, K. J., & Smith, A. B. (2012). Resolving phylogenetic signal from noise when divergence is rapid: A new look at the old problem of echinoderm class relationships. Molecular Phylogenetics and Evolution, 62(1), 27–34. doi:10.1016/j.ympev.2011.08.028
-
Sperling, E. A., Pisani, D., & Peterson, K. J. (2011). Molecular paleobiological insights into the origin of the Brachiopoda. Evolution & Development, 13(3), 290–303. doi:10.1111/j.1525-142×.2011.00480.x
-
Tarver, J. E., Donoghue, P. C. J., & Peterson, K. J. (2012). Do miRNAs have a deep evolutionary history?. BioEssays, 34(10), 857–866. doi:10.1002/bies.201200055
-
Vinther, J., Sperling, E. A., Briggs, D. E. G., & Peterson, K. J. (2011). A molecular palaeobiological hypothesis for the origin of aplacophoran molluscs and their derivation from chiton-like ancestors. Proceedings of the Royal Society B: Biological Sciences, 279(1732), 1259–1268. doi:10.1098/rspb.2011.1773
-
Wiegmann, B. M., Trautwein, M. D., Winkler, I. S., Barr, N. B., Kim, J-W., Lambkin, C., … Yeates, D. K. (2011). Episodic radiations in the fly tree of life. Proceedings of the National Academy of Sciences, 108(14), 5690–5695. doi:10.1073/pnas.1012675108
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Kevin Peterson
Project Investigator
-
RELATED OBJECTIVES:
Objective 4.2
Production of complex life.