2012 Annual Science Report
 Georgia Institute of Technology
Reporting | SEP 2011 – AUG 2012
Georgia Institute of Technology
Reporting | SEP 2011 – AUG 2012
Experimental Evolution and Genomic Analysis of an E. Coli Containing a Resurrected Ancestral Gene
Project Summary
We have previously described a paleo-experimental evolution system that combines Ancestral Sequence Reconstruction (ASR) with experimental evolution in the laboratory. Briefly, we designed a system that is composed of an organism with a short generation time and a protein under strong selective constraints in the modern host but whose ancestral genotype and phenotype, if genomically integrated, causes the modern host to be less fit than a modern population hosting the modern form of the protein. The modern organism hosting the resurrected protein would obviously need to be viable, but sick. E. coli and a protein, whose ancestral sequences are available termed Elongation Factor-Tu (EF-Tu), turned out to be ideal for this type of experiment.
Project Progress
To exploit a system that allows us to investigate the stages of adaptation and to address aims concerned with natural history and molecular evolution, we replaced endogenous E. coli EF-Tu with a 500mya ancestral EF-Tu. Initial experiments showed that replacing the modern bacterial EF-Tu gene with its ~500 million year old ancestor extends the bacterial doubling time by two-fold. We then measured the immediate phenotypic effects of these insertions/deletions on the bacteria via fitness competition experiments. Such measurements showed a ~25% decrease in the relative fitness (W=0.75) relative to the bacteria that contains modern EF-Tu (W=1.0). To examine the subsequent evolution of these 'sick’ modern E.coli harboring resurrected genes, we setup an experimental evolution system in 8-parallel lineages originated from 8-independent clones in Davis minimal medium for 2000 generations. We followed the fitness change in 8-parallel lineages in every 500 generations by competitions between ancestral strains harboring the ancestral EF-Tu against the E.coli strain harboring both of the wild type EF-Tu. In less than 500 generations of evolution, populations reach back to fitness value comparable to that of E.coli harboring a modern EF, further we measured an increase of ~%35 (Wave=1.1) for 8 lineages in less than 2000 generations.
To provide further insights into how evolution operates at the systems level, we coupled this paleo-experimental evolution system with whole genome sequencing. Sequencing the genomes of 8-_E.coli_ populations sampled every 500 generations allowed us to delineate the mutations that have potentially accumulated in the ancient-EF and the rest of the bacterial genome during experimental evolution. The unevolved ancestral strain REL606 (GenBank accession number NC_012967.1) served as a reference for identifying the mutations in the evolved lineages. Even though no beneficial mutation accumulated in the ancient EF region, we detected an average of 2 non-synonymous mutations per lineage, per generation. We also observed that several of these proteins accumulate mutations in more than one lineage, suggesting a strong evidence for parallel evolution. Interestingly, most of the non-synonymous mutations that are accumulated in these replicate populations are either directly interacting or are within the interacting network of EF-Tu. We are currently investigating the role of these mutations both in terms of their fitness benefit and in terms of shaping the population’s evolutionary trajectories. Further, we are observing how these mutations affected the evolution of the whole protein interaction network of EF-Tu by biochemically identifying the effect of these mutations on protein-protein interactions.
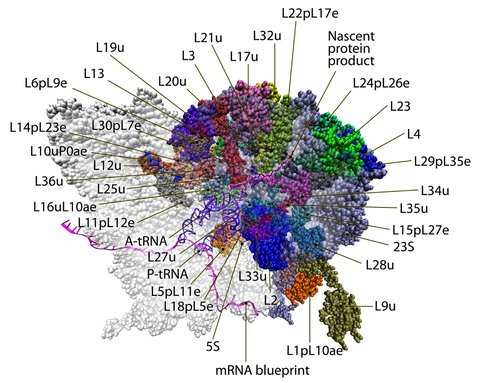
The E .coli 70S ribosome. Portions of the rRNA are hidden to emphasize the rPRoteins, active site, and nascent peptide. The nascent product is ~5.5 nm long. We show the small subunit in white. This image combines renderings of aligned models from several labs (Llano-Sotelo et al., 2010; Agirrezabala et al., 2012; Frauenfeld et al., 2011; Bingel-Erlenmeyer et al., 2008; Zhou et al., 2012; Seidelt et al., 2009; Schmeing et al., 2002) (retrieved from DARC)
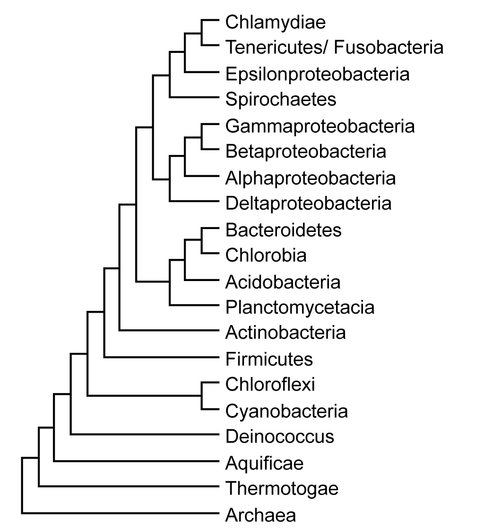
A model of bacterial evolution inferred using Bayesian inference (Ronquist et al., 2012).
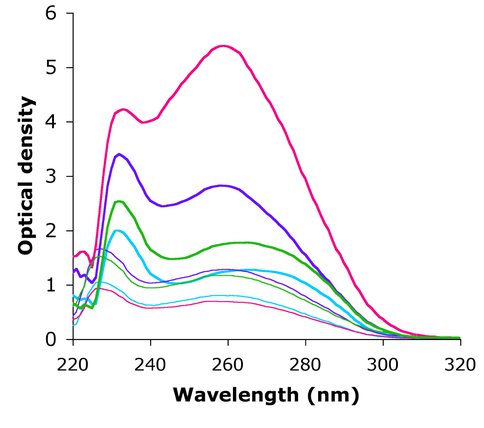
Absorbance spectra of wash and elution fractions for preliminary ribosome purifications. Pink = L12-HisTag, Purple = Ancestral-L12-HisTag, Green = Ancestral-L2-HisTag, Blue = Empty plasmid vector pET21a. Thin lines are spectra for wash fractions and thick lines are for elution fractions.
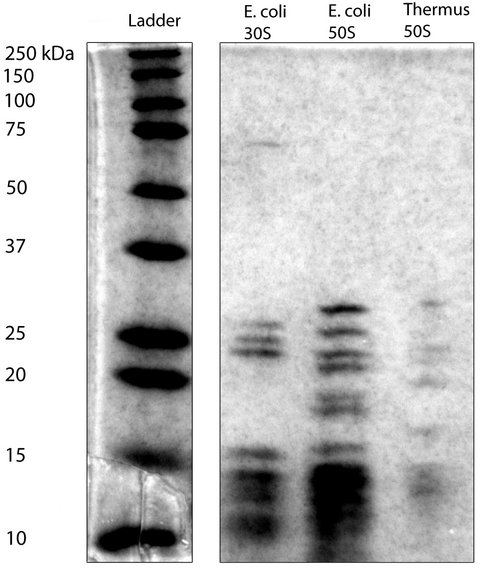
SDS gel stained with Coomassie
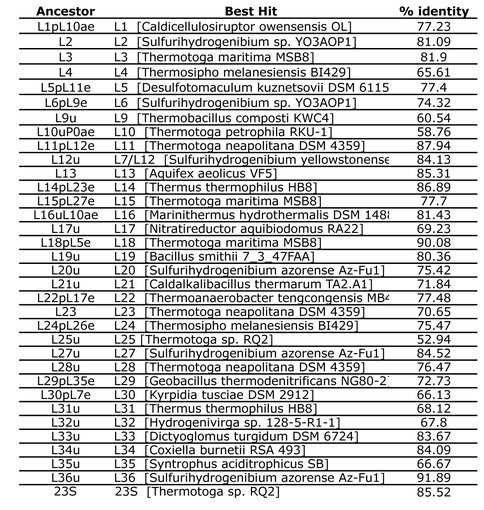
After inferring ancestral 50S ribosomal sequences, we performed NCBI BLAST searches using the ancestral predictions as queries. Listed are the top hits for each search as well as the % identity of a pairwise alignment between query and subject.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Betül Kacar
Research Staff
Joshua Stern
Graduate Student
-
RELATED OBJECTIVES:
Objective 3.4
Origins of cellularity and protobiological systems
Objective 4.1
Earth's early biosphere.