2012 Annual Science Report
 Georgia Institute of Technology
Reporting | SEP 2011 – AUG 2012
Georgia Institute of Technology
Reporting | SEP 2011 – AUG 2012
Deconstruction of the Ribosome
Project Summary
In the ribosome, RNA and protein are fully interdependent, part of the highly complex system of translation. We demonstrate here that isolated Domain III of 23S rRNA from Thermus thermophilus retains function after being split essentially in half. Chemical footprinting shows that a core of Domain III (DIIIcore), obtained replacement of helices 54-59 with a simple stem-loop, folds to a near-native state in the presence of Mg2+ ions. Both DIII and DIIIcore form specific complexes with ribosomal protein L23 in vivo, as indicated with a yeast three-hybrid experiment. L23 has a globular domain on the LSU surface and an extension (L23peptide) that penetrates into the ribosomal large subunit. In the assembled LSU, L23peptide (amino acids 58-79) traverses the surface of DIIIcore. In solution, DIIIcore forms a stable 1:1 complex with L23peptide, as observed with spectroscopic assay. The experiments described here are intended to recapitulate steps in early ribosomal evolution. We have previously proposed that some of the extensions of ribosomal proteins are molecular fossils that predate the globular protein domains in evolution. In our favored model of ribosomal origins, small independently-folding RNA elements associated with short peptides. Such complexes assembled to form a primitive peptidyl transferase center. The PTC evolved into the modern LSU, in a series of cooptions that left unaltered the basic structure and function of the PTC. This model predicts a continuous size distribution of folding and assembly elements within the LSU. We anticipate autonomy and specificity of folding and interaction of small, mid-sized and large rRNA and protein components.
Project Progress
The origin of complexity in biological systems is a classical evolutionary dilemma. On a molecular level, translation is perhaps the most complex of biological processes. The ribosome is responsible for protein synthesis in all organisms. The two ribosomal subunits, the small subunit (SSU) and the large subunit (LSU), come together during translation to form the active ribosome. While ribosomal RNAs (rRNAs) perform the crucial activities of decoding and peptidyl transfer (1-3), proteins charge tRNAs (4), stabilize inter- and intra-subunit ribosomal interactions and assist in ribosomal assembly (5,6).
Translation is one of our most direct macromolecular connections to early life (7-13). Comparative genomic reconstructions suggest that the core of the translation system was mature and fully functional in the last common universal ancestor (14,15). The LSU in bacteria and archaea contains a 23S rRNA and a much smaller 5S rRNA. Although the 23S rRNA is generally described as containing six 2° domains of rRNA (Figure 1A) (16), the 3D architecture appears monolithic (1,17,18). Recent experimental evidence demonstrates that Domain III of the LSU folds autonomously into a near-native 3D structure (19).
In translation, RNA and protein are fully interdependent in a system of nearly overwhelming molecular complexity. An elaborate and multi-step process of indirect templating is imposed by fundamental differences in the structures of RNA compared to protein. However, first principles require that origins and evolution of RNA/protein interdependence in the ribosome followed a sequence of incremental and elementary steps, each linked to immediate fitness, without foresight. It seems reasonable to propose that the ribosome can be understood and explained in part by recapitulating these elementary steps (11).
Here we experimentally recapitulate steps in early ribosomal evolution by deconstructing the ribosome. We demonstrate that Domain III of Thermus thermophilus 23S rRNA (DIII, Figure 1) can retain function after being split essentially in half. A core element of Domain III (DIIIcore, Figure 1), obtained by the elimination of helices 54-59, retains the ability to fold to a near-native state. Both DIII and with DIIIcore form specific complexes with ribosomal protein L23, as observed in vivo with a yeast three-hybrid method. L23 has a globular domain on the LSU surface and an extension (L23peptide) that penetrates into the LSU (20). L23peptide (amino acids 58-79 from T. thermophilus L23) traverses the surface of DIIIcore (Figure 2). In solution, DIIIcore forms a specific 1:1 complex with L23peptide, as observed with spectroscopic assay. The interactions of fragmented ribosomal components are predicted by interactions observed in the intact LSU (Figure 2).
Domain III rRNA forms part of the exit tunnel in the assembled ribosome. The exit tunnel is also formed by four evolutionarily conserved ribosomal proteins (L22, L23, L24 and L29) as well as other kingdom-specific proteins. Ribosomal protein L23 is unique because it functions as a docking site for both trigger factor (TF) and signal recognition particle (SRP) (21).
I. Molecular interactions within DIII.
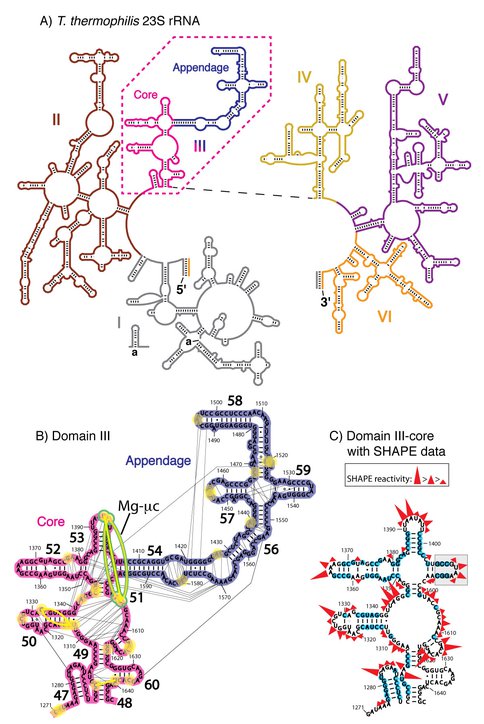
rRNA-rRNA interactions. The pattern of molecular interactions within DIII of the LSU crystal structure T. thermophilus, PDB entry 2J01 (3)] suggests that DIII contains an independent subdomain, DIIIcore, composed of nucleotides 1271-1406 and 1596-1646. The independent integrity of DIIIcore is supported by a large number of intra-subdomain tertiary interactions. Intra-subdomain interactions are both RNA-RNA and RNA-Mg2+-RNA interactions that include base pairing, base stacking, phosphate-base, phosphate-sugar, as well as phosphate-Mg2+-phosphate interactions. Few interactions were observed between DIIIcore and DIIIH54-59 (Figure 1B and Table S1).
Magnesium interactions. Mg2+ plays a critical role in the folding and assembly of large RNAs (22,23). The bacterial and archaeal LSUs are rich in Mg2+ ions (24,25) and are stabilized by an RNA-Mg2+ motif called the Mg2+-microcluster (Mg-µc) (26,27), defined by: (i) two proximal Mg2+ ions chelated by a common bridging phosphate group in the form Mg(a) – O1P – P – O2P – Mg(b); (ii) a 10-membered chelation ring that includes the bridging phosphate in the form Mg(a) – OP – P – O5′ – C5′ – C4′ – C3′ – O3′ – P – OP – Mg(a); (iii) octahedral coordination of both Mg2+ ions by a combination of water molecules and RNA phosphate groups; and (iv) nucleotides in non-canonical RNA conformations and unstacked bases. A single Mg-µc is observed in DIII, which is contained entirely within DIIIcore. The two Mg2+ ions of this Mg-µc form a tight interaction network with RNA nucleotides of DIIIcore, and do not interact with RNA outside of DIIIcore. This Mg-µc involves nucleotides A1395, A1603 and C1604, bridging helices 51 and 53 of DIIIcore (Figure 1B).
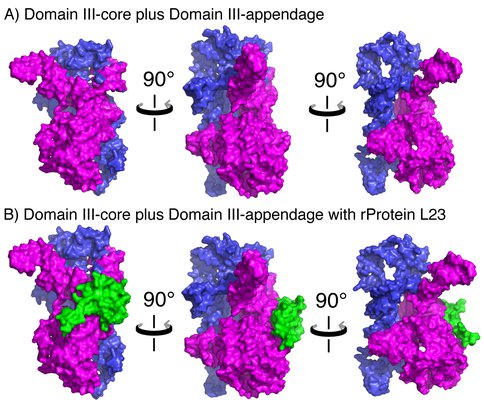
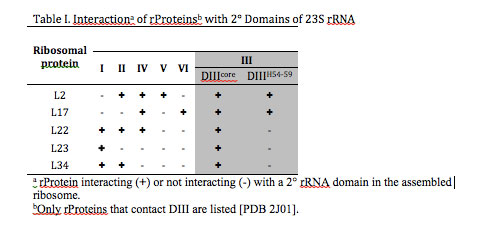
rRNA-rProtein interactions. Based on the proximity of atoms within the assembled LSU, rProteins L2, L17, L22, L23, and L34 interact with DIII rRNA. Contacts were defined as a DIII atom – rProtein atom pair in which the distance between the atoms was less than the sum of their Van der Waals radii. L2, L17, L22, and L34 are bound in rRNA pockets formed partially by DIII, bridging DIII and at least two other 2º domains. By contrast, L23 interacts only with Domains I and III (Table I). L23-DIII contacts are fully contained within DIIIcore.
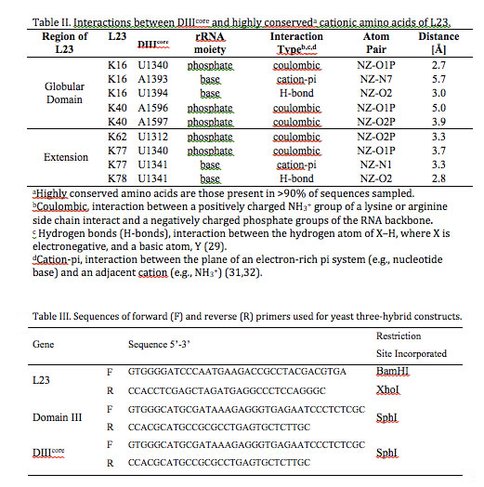
DIIIcore interactions with L23. The amino acids of L23 that interact with DIIIcore are highly conserved over phylogeny. Sequence alignments of L23 from a subset of 121 organisms representing the three domains of life (28) reveal five cationic amino acids K16, K40, K62, K77, and K78 that are highly conserved (present in >90% of the sequences sampled). All conserved amino acids of L23 interact exclusively with DIIIcore, suggesting a deep evolutionary relationship.
With geometric analysis, we have parsed the types of molecular interactions (coulombic, cation-pi, and hydrogen bond) that link DIIIcore and L23 (Table II). L23 primarily interacts coulombically with DIIIcore: the positively charged functional groups of lysines and arginines interacts with negatively charged phosphate groups of the rRNA. These electrostatic interactions (defined here by a cutoff distance of 5.5 Å) are observed for conserved amino acids K16, K40, K62, and K77 of L23. Hydrogen bonds (H-bonds) are an attractive interaction between the hydrogen atom of X–H, where X is electronegative, and a basic atom, Y (29). H-bonds are defined by a cutoff distance of 3.4 Å between atoms X and Y (Table II). L23 interacts with DIIIcore bases via H-bonding of K16 to U1340 and K78 to U1341. Two cation-pi interactions (30-32) are observed between cationic amino acids of L23 and the aromatic systems of bases in DIIIcore: K16 interacts with A1393 and K77 interacts with U1341. Cation-pi interactions occur between the plane of an electron-rich pi system (e.g., nucleotide base) and an adjacent cation (e.g., NH3+). Cation-pi interactions are defined by a cutoff distance of 6 Å and a cutoff angle θ of 60o between the normal to the plane of the pi-system and the vector connecting the center of the pi-system of a nucleobase to the NH3+ of an amino acid R-group.
II. Secondary structure of DIIIcore
DIIIcore is composed of two fragments of rRNA (Figure 1B). To reform DIIIcore as a single continuous RNA polymer, we joined the two fragments together with a stem-loop. This addition is unlikely to perturb the structure since the termini of the two DIIIcore fragments are opposed in a double-stranded A-form helical arm that can directly dock onto the stem-loop (Figure 1C). The stem-loop used here (5’-gccGUAAggc-3’), is a GNRA stem-loop (33) on a three base pair helical stem (lower case letters).
The DIIIcore autonomously forms LSU-like secondary structure. We used SHAPE (34,35) to probe the structure of DIIIcore. SHAPE exploits the reactivity of the 2′-hydroxyl groups of RNA with electrophilic chemical probing reagents such as NMIA (N-methylisatoic anhydride). The relative reactivities of the 2′-hydroxyl groups of a RNA are dependent primarily on local RNA flexibility. Consequently, paired nucleotides within helical regions are generally less flexible and less reactive with SHAPE reagents than unpaired nucleotides. Local RNA flexibility, and therefore SHAPE reactivity, is also altered by tertiary interactions facilitated by Mg2+ ions. We previously used SHAPE to demonstrate that the secondary structure and Mg2+-dependent folding of DIII are retained when DIII is excised from the 23S rRNA (36).
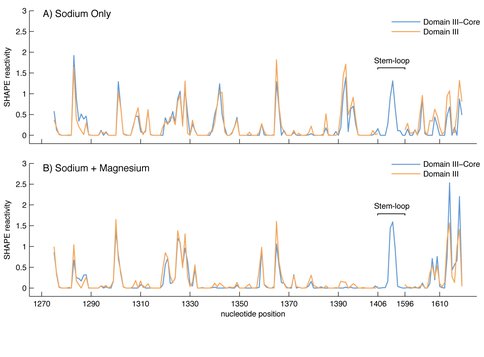
The SHAPE reactivity of DIIIcore was initially determined in the absence of Mg2+ (Figures 1C and 3A). Under these conditions, the RNA is expected to form secondary structure but not a collapsed state with long-range tertiary interactions (37,38). Nucleotides of DIIIcore were ranked using their absolute SHAPE reactivities relative to A1365 (which had highest SHAPE reactivity). The nucleotides were then binned into four groups, as indicated in Figure 1C [** LDW I don’t see this, there is no Figure 1C]. SHAPE reactivity data obtained here for DIIIcore corresponds very closely with its canonical secondary structure within the 23S rRNA. More importantly, as illustrated by the overlaid SHAPE traces (Figure 3), the reactivities of DIII and DIIIcore are essentially identical along the length of the DIIIcore sequence, except where the stem-loop was inserted to connect the two fragments of DIIIcore. The high degree of similarity in SHAPE reactivities suggests that the secondary structure of DIIIcore alone is the same as DIII in the absence of Mg2+.
In the presence of Mg2+ ions, DIIIcore autonomously forms a compact near-native structure. The folding of large RNAs, from secondary structure to collapsed native states with long-range tertiary interactions, is Mg2+-dependent (22,37,38). The native state of DIII rRNA, as inferred from the 3D structure of the assembled LSU, is stabilized by extensive networks of intra-domain tertiary base-base, base-backbone and backbone-Mg2+-backbone interactions (including an Mg-µc). Consistent with Mg2+-induced collapse to form a near-native state, the changes in SHAPE reactivity of DIII upon addition of Mg2+ are widely dispersed over DIII rRNA (Figure 3B). SHAPE reactivities increase at some sites and decrease at others. This pattern of Mg2+-dependent SHAPE reactivity reflects (i) specific Mg2+ binding to RNA, (ii) diffuse interactions of Mg2+ with the RNA, and/or (iii) formation of rRNA-rRNA tertiary interactions. Mg2+-dependent SHAPE reactivity has been demonstrated previously for tRNA (35), RNase P (39), and DIII (36). The effects of Mg2+ on the SHAPE reactivities of DIIIcore and DIII are very similar (Figure 3B). The Mg2+-dependence of DIIIcore folding is therefore retained when DIIIcore is excised from DIII.
Some of the most intense Mg2+-induced changes in SHAPE reactivity of DIIIcore reflect direct Mg2+ interactions with the rRNA. The SHAPE reactivities of 20 nucleotides change by 50% or more upon the addition of Mg2+. Nineteen of these nucleotides interact directly with Mg2+ or are in the close proximity to Mg2+ in the 3D structure. For example, among nucleotides A1395, A1603 and C1604, which form a Mg2+-μc motif, A1395 and A1603 report changes in SHAPE reactivity of >80% upon addition of magnesium. Other nucleotides directly involved in Mg2+ interactions in the crystal structure that give large changes in SHAPE reactivity upon addition of Mg2+ are U1313, A1342, A1614 and C1615. Nucleotides G1283, A1284, U1300, U1341, G1343, A1392, A1393, U1396, C1607, A1609, C1617 and A1618 give large Mg2+-induced changes in SHAPE reactivity, and are in close proximity to Mg2+ ions.
III. RNA-Protein Interactions.
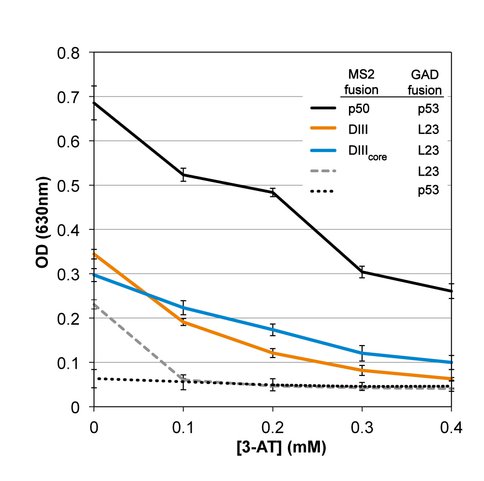
DIII and DIIIcore interact with L23 in vivo. We used the yeast three-hybrid system (40-42) to assay the interaction of DIII or DIIIcore with intact L23 in vivo (Figure 4). An interaction of RNA with protein in yeast results in resistance to 3-amino-1,2,4-triazole (3-AT) due to increased expression of reporter gene HIS3 (42).
In the yeast three-hybrid in vivo assay, both DIII and DIIIcore interact with L23 (Figure 5). In vivo interaction of each RNA with L23 resulted in similar expression of reporter gene HIS3, as determined by a 3-AT resistance of approximately 0.4 mM. Although expression is less than that observed for a positive control (p50 RNA/p53 protein), it is well above background (assay without DIII or DIIIcore), confirming in vivo RNA-protein binding.
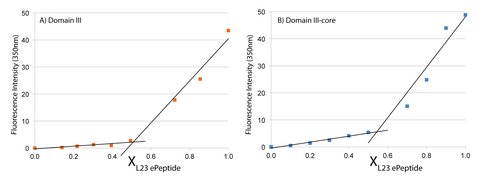
L22peptide forms a 1:1 complex with DIII and with DIIIcore in vitro. L23peptide penetrates into the LSU and interacts with DIIIcore. We characterized the binding and stoichiometry of the interaction of L23peptide with DIII or DIIIcore in vitro using the method of continuous variation (43,44). A series of solutions with constant ratios. To enable detection by fluorescence, we added an intrinsic fluorophore (Trp) to the C-terminal end of the L23peptide. This amino acid is a tryptophan in the E. coli L23, but not in the T. thermophilus L23. We inferred the stoichiometry of binding from the discontinuity in the plot of fluorescence intensity versus mole fraction of L23peptide. The results indicate that L23peptide forms a 1:1 complex with both DIII and with DIIIcore (Figure 6). These experiments were conducted in the presence of monovalent cations only. Upon the addition of Mg2+, a more complex interaction between L23peptide and both DIII and DIIIcore is observed (work in progress).
The architecture of the ribosome has profound implications for our understanding of ribosomal assembly and function, and evolution of early life. The architecture of the ribosome can be probed experimentally. Both rRNA and rProteins can be fragmented into hypothetical evolutionary and functional units and the units can be assayed for ability to fold and function.
Many rProteins have globular domains on the subunit surface, and extensions that penetrate the subunit core (20). We have proposed that some of the these extensions of rProteins are molecular fossils that predate the globular protein domains in evolution (8). In our conception of ribosomal origins, small independently-folding RNA elements associated with short peptides. Such complexes assembled to form a primitive peptidyl transferase center, which performed non-coded condensation reactions, without participation of the SSU [also see Fox, (13)]. The PTC evolved into the modern LSU, in a series of cooptions that left unaltered the basic structure and function of the PTC. This model predicts a continuous size distribution of folding and assembly elements within the LSU. We anticipate autonomy and specificity of folding and interaction of small, mid-sized and large rRNA and protein components.
Here we have deconstructed a portion of the ribosome, isolating relatively small RNA and protein elements that retain the ability to fold and assemble. We have extracted Domain III from the 23S rRNA and further reduced Domain III to Domain IIIcore. DIIIcore is defined by G1271-U1406 and A1596-G1647). Helices 54-59 (DIIIH54-59, C1407-G1595) were eliminated from Domain III to form DIIIcore (Figure 1). Analysis of molecular interactions within T. thermophilus crystal structure (PDB 2J01) (3) reveals that many tertiary interactions stabilize DIIIcore. Few tertiary interactions are observed between DIIIcore and DIIIH54-59 or within DIIIH54-59. DIIIcore is stabilized by both rRNA-rRNA interactions and RNA-Mg2+-RNA interactions.
Domain IIIcore retains the ability to fold into a near native structure. rProtein L23 specifically interacts in vivo with both DIII and DIIIcore, independently of other components of the ribosome. This interaction suggests that DIIIcore represents a functional rRNA unit in the context of DIII-L23 assembly. Furthermore, the ‘un-structured’ extension of rProtein L23 that we call L23peptide sustains the interaction function in vitro, with both DIII and DIIIcore. The L23 extension peptide supports assembly independently of any stabilizing effects contributed by the globular domain of L23. The ability of L23peptide to form a 1:1 complex with both DIII and DIIIcore suggests that L23peptide is a functional rProtein unit.
The results here are consistent with our model of continuous size distribution of folding and assembly elements. We demonstrate that a short fragment of a ribosomal proteins (rProtein extension L23peptide; 21 aa long), does bind specifically to fragments of ribosomal RNA (DIIIcore and DIII) in vitro.
The biological functions of ribosomal protein extensions are not always obvious. The extensions of rProteins L4 and L22 can be deleted from E. coli without deleterious consequence in ribosomal assembly and function (45). Mutations in the extensions of L4 and L22 confer macrolide antibiotic resistance in E. coli (46). Little is known about the requirement for the L23 extension, which lines the exit tunnel; however, a recent study suggests that the L23 extension could interact with ribosome-bound nascent polypeptides, triggering a conformational change in L23 globular domain, where TF binds, to regulate TF recruitment (47).
Domain III of the 23S is phylogenetically variable and, in some mitochondrial rRNAs, is absent (48). Sequence and structure corresponding with DIIIH54-59 is particularly variable (49). In fragmented bacterial 23S rRNAs, Domain III contains unique fragmentation site that lacks an stem-loop structure found at other fragmentation sites (50). rProtein L23 is essential in E. coli (51), but expendable in Bacillus subtilis (52). In addition to functions in docking of both TF and SRP, L23 is capable of binding to SecA, which is proposed to post-translationally recognize secretory proteins (53). L23 is not present in eukaryotic ribosomes; however, rProtein L39e occupies a similar position in the eukaryotic and archaeal exit tunnel (54).
The ribosome as whole is quite robust and has functional plasticity. L23 is multi-functional yet is not universally necessary for function; similarly, Domain III is highly variable and not universally required for translation. It is intriguing that L23 maintains in vivo binding to DIII and DIIIcore rRNA in the absence of supporting interactions from other rRNA and rProteins in the assembled ribosome. Even further, lacking supporting interactions, L23peptide maintains 1:1 in vitro binding stoichiometry with DIIIcore despite the flexibility of the peptide extension in the exit tunnel.
Our work supports a platform on which deconstructing the ribosome and exploring its units of modification further elucidates structure, function and evolution of this complex molecular machine. In addition, the non-uniform distribution of the DIII-L23 interaction unit offers targets for antibiotics. The ability to deconstruct the DIII-L23 interaction from the fully assembled ribosome both in vivo and in vitro also offers relatively simple, and potentially high-throughput, assays to test antibiotic activity.
References.
1. Ban, N., Nissen, P., Hansen, J., Moore, P.B. and Steitz, T.A. (2000) The Complete Atomic Structure of the Large Ribosomal Subunit at 2.4 Å Resolution. Science (New York, N.Y.), 289, 905-920.
2. Noller, H.F., Hoffarth, V. and Zimniak, L. (1992) Unusual Resistance of Peptidyl Transferase to Protein Extraction Procedures. Science (New York, N.Y.), 256, 1416-1419.
3. Selmer, M., Dunham, C.M., Murphy, F.V., Weixlbaumer, A., Petry, S., Kelley, A.C., Weir, J.R. and Ramakrishnan, V. (2006) Structure of the 70S Ribosome Complexed with mRNA and tRNA. Science (New York, N.Y.), 313, 1935-1942.
4. Schimmel, P. (2008) Development of tRNA Synthetases and Connection to Genetic Code and Disease. Protein Sci., 17, 1643-1652.
5. Klein, D.J., Moore, P.B. and Steitz, T.A. (2004) The Roles of Ribosomal Proteins in the Structure Assembly, and Evolution of the Large Ribosomal Subunit. J. Mol. Biol., 340, 141-177.
6. Rohl, R. and Nierhaus, K.H. (1982) Assembly Map of the Large Subunit (50S) of Escherichia Coli Ribosomes. Proc. Natl. Acad. Sci. U. S. A.. 79, 729-733.
7. Belousoff, M.J., Davidovich, C., Zimmerman, E., Caspi, Y., Wekselman, I., Rozenszajn, L., Shapira, T., Sade-Falk, O., Taha, L., Bashan, A. et al. (2010) Ancient Machinery Embedded in the Contemporary Ribosome. Biochem. Soc. Trans., 38, 422-427.
8. Hsiao, C., Mohan, S., Kalahar, B.K. and Williams, L.D. (2009) Peeling the Onion: Ribosomes Are Ancient Molecular Fossils. Mol. Biol. Evol., 26, 2415-2425.
9. Bokov, K. and Steinberg, S.V. (2009) A Hierarchical Model for Evolution of 23S Ribosomal RNA. Nature, 457, 977-980.
10. Smith, T.F., Lee, J.C., Gutell, R.R. and Hartman, H. (2008) The Origin and Evolution of the Ribosome. Biol. Direct, 3, 16.
11. Wolf, Y.I. and Koonin, E.V. (2007) On the Origin of the Translation System and the Genetic Code in the RNA World by Means of Natural Selection, Exaptation, and Subfunctionalization. Biol. Direct, 2, 14.
12. Woese, C.R. (2001) Translation: In Retrospect and Prospect. RNA, 78, 1055-1067.
13. Fox, G.E. (2010) Origin and Evolution of the Ribosome. Cold Spring Harb. Perspect. Biol., 2, a003483.
14. Anantharaman, V., Koonin, E.V. and Aravind, L. (2002) Comparative Genomics and Evolution of Proteins Involved in RNA Metabolism. Nucleic Acids Res., 30, 1427-1464.
15. Olsen, G.J. and Woese, C.R. (1993) Ribosomal RNA: A Key to Phylogeny. FASEB J., 7, 113-123.
16. Noller, H.F., Kop, J., Wheaton, V., Brosius, J., Gutell, R.R., Kopylov, A.M., Dohme, F., Herr, W., Stahl, D.A., Gupta, R. et al. (1981) Secondary Structure Model for 23S Ribosomal RNA. Nucleic Acids Res., 9, 6167-6189.
17. Tumminia, S.J., Hellmann, W., Wall, J.S. and Boublik, M. (1994) Visualization of Protein-Nucleic Acid Interactions Involved in the in Vitro Assembly of the Escherichia Coli 50S Ribosomal Subunit. J. Mol. Biol., 235, 1239-1250.
18. Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H. and Noller, H.F. (2001) Crystal Structure of the Ribosome at 5.5 Å Resolution. Science (New York, N.Y.), 292, 883-896.
19. Athavale, S.S., Gossett, J.J., Hsiao, C., Bowman, J.C., O’Neill, E., Hershkovitz, E., Preeprem, T., Hud, N.V., Wartell, R.M., Harvey, S.C. et al. (2012) Domain III of the T. thermophilus 23S rRNA Folds Independently to a near-Native State. RNA, 18, 752-758.
20. Wilson, D.N. and Nierhaus, K.H. (2005) Ribosomal Proteins in the Spotlight. Crit. Rev. Biochem. Mol. Biol., 40, 243-267.
21. Baneyx, F. (2012) A Ribosomal Surprise. Biotechnol. J., 7, 326-327.
22. Bowman, J.C., Lenz, T.K., Hud, N.V. and Williams, L.D. (2012) Cations in Charge: Magnesium Ions in RNA Folding and Catalysis Curr. Opin. Struct. Biol., 22, 262-272.
23. Draper, D.E., Grilley, D. and Soto, A.M. (2005) Ions and RNA Folding. Annu. Rev. Biophys. Biomol. Struct., 34, 221-243.
24. Klein, D.J., Moore, P.B. and Steitz, T.A. (2004) The Contribution of Metal Ions to the Structural Stability of the Large Ribosomal Subunit. RNA, 10, 1366-1379.
25. Hsiao, C., Tannenbaum, M., VanDeusen, H., Hershkovitz, E., Perng, G., Tannenbaum, A. and Williams, L.D. (2008) In Hud, N. (ed.), Nucleic Acid Metal Ion Interactions. The Royal Society of Chemistry, London, pp. 1-35.
26. Hsiao, C. and Williams, L.D. (2009) A Recurrent Magnesium-Binding Motif Provides a Framework for the Ribosomal Peptidyl Transferase Center. Nucleic Acids Res., 37, 3134-3142.
27. Petrov, A.S., Bowman, J.C., Harvey, S.C. and Williams, L.D. (2011) Bidentate RNA-Magnesium Clamps: On the Origin of the Special Role of Magnesium in RNA Folding. RNA, 17, 291-297.
28. Petrov, A., Bernier, C., Hsiao, C., Okafor, C.D., Tannenbaum, E., Stern, J., Gaucher, E., Schneider, D.M., Harvey, S.C. and Williams, L.D. (2012) RNA-Magnesium-Protein Interactions in Large Ribosomal Subunit. J. Phys. Chem. B, 116, 8113-8120.
29. Jeffrey, G.A. (1997) An Introduction to Hydrogen Bonding. Oxford University Press, New York.
30. Gallivan, J.P. and Dougherty, D.A. (1999) Cation-Pi Interactions in Structural Biology. Proc. Natl. Acad. Sci. U. S. A., 96, 9459-9464.
31. Dougherty, D.A. (1997) Electrostatic Models for the Cation-Pi Interaction and Related Non-Covalent Interactions. J. Am. Chem. Soc., 214, 166.
32. Ma, J.C. and Dougherty, D.A. (1997) The Cation-Pi Interaction. Chem. Rev., 97, 1303-1324.
33. Mohan, S., Hsiao, C., Bowman, J.C., Wartell, R. and Williams, L.D. (2010) RNA Tetraloop Folding Reveals Tension between Backbone Restraints and Molecular Interactions. J. Am. Chem. Soc., 132, 12679-12689.
34. Merino, E.J., Wilkinson, K.A., Coughlan, J.L. and Weeks, K.M. (2005) RNA Structure Analysis at Single Nucleotide Resolution by Selective 2’-Hydroxyl Acylation and Primer Extension (SHAPE). J. Am. Chem. Soc., 127, 4223-4231.
35. Wilkinson, K.A., Merino, E.J. and Weeks, K.M. (2005) RNA SHAPE Chemistry Reveals Nonhierarchical Interactions Dominate Equilibrium Structural Transitions in tRNA(Asp) Transcripts. J. Am. Chem. Soc., 127, 4659-4667.
36. Athavale, S.S., Petrov, A.S., Hsiao, C., Watkins, D., Prickett, C.D., Gossett, J.J., Lie, L., Bowman, J.C., O’Neill, E., Bernier, C.R. et al. (2012) RNA Folding and Catalysis Mediated by Iron (II). PLoS ONE, 7, e38024.
37. Brion, P. and Westhof, E. (1997) Hierarchy and Dynamics of RNA Folding. Annu. Rev. Biophys. Biomol. Struct., 26, 113-137.
38. Draper, D.E. (2008) RNA Folding: Thermodynamic and Molecular Descriptions of the Roles of Ions. Biophys. J., 95, 5489-5495.
39. Mortimer, S.A. and Weeks, K.M. (2008) Time-Resolved RNA SHAPE Chemistry. J. Am. Chem. Soc., 130, 16178-16180.
40. Bernstein, D.S., Buter, N., Stumpf, C. and Wickens, M. (2002) Analyzing mRNA-Protein Complexes Using a Yeast Three-Hybrid System. Methods (San Diego, Calif.), 26, 123-141.
41. Hook, B., Bernstein, D., Zhang, B. and Wickens, M. (2005) RNA-Protein Interactions in the Yeast Three-Hybrid System: Affinity, Sensitivity, and Enhanced Library Screening. RNA, 11, 227-233.
42. Stumpf, C.R., Opperman, L. and Wickens, M. (2008) Analysis of RNA-Protein Interactions Using a Yeast Three-Hybrid System. Methods Enzymol., 449, 295-315.
43. Job, P. (1928) Studies on the Formation of Complex Minerals in Solution and on Their Stability. Annales De Chimie France, 9, 113-203.
44. Cantor, C. and Schimmel, P. (1984) Biophysical Chemistry (I-III). Academic Press, New York.
45. Zengel, J.M., Jerauld, A., Walker, A., Wahl, M.C. and Lindahl, L. (2003) The Extended Loops of Ribosomal Proteins L4 and L22 Are Not Required for Ribosome Assembly or L4-Mediated Autogenous Control. RNA, 9, 1188-1197.
46. Diner, E.J. and Hayes, C.S. (2009) Recombineering Reveals a Diverse Collection of Ribosomal Proteins L4 and L22 That Confer Resistance to Macrolide Antibiotics. J. Mol. Biol., 386, 300-315.
47. Lin, K.F., Sun, C.S., Huang, Y.C., Chan, S.I., Koubek, J., Wu, T.H. and Huang, J.J. (2012) Cotranslational Protein Folding within the Ribosome Tunnel Influences Trigger-Factor Recruitment. Biophys. J., 102, 2818-2827.
48. Mears, J.A., Cannone, J.J., Stagg, S.M., Gutell, R.R., Agrawal, R.K. and Harvey, S.C. (2002) Modeling a Minimal Ribosome Based on Comparative Sequence Analysis. J. Mol. Biol., 321, 215-234.
49. Van Camp, G., Chapelle, S. and De Wachter, R. (1993) Amplification and Sequencing of Variable Regions in Bacterial 23S Ribosomal RNA Genes with Conserved Primer Sequences. Curr. Microbiol., 27, 147-151.
50. Evguenieva-Hackenberg, E. (2005) Bacterial Ribosomal RNA in Pieces. Mol. Microbiol., 57, 318-325.
51. Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., Datsenko, K.A., Tomita, M., Wanner, B.L. and Mori, H. (2006) Construction of Escherichia Coli K-12 in-Frame, Single-Gene Knockout Mutants: The Keio Collection. Mol. Syst. Biol., 2.
52. Akanuma, G., Nanamiya, H., Natori, Y., Yano, K., Suzuki, S., Omata, S., Ishizuka, M., Sekine, Y. and Kawamura, F. (2012) Inactivation of Ribosomal Protein Genes in Bacillus Subtilis Reveals Importance of Each Ribosomal Protein for Cell Proliferation and Cell Differentiation. J. Bacteriol., 194, 6282-6291.
53. Huber, D., Rajagopalan, N., Preissler, S., Rocco, M.A., Merz, F., Kramer, G. and Bukau, B. (2011) Seca Interacts with Ribosomes in Order to Facilitate Posttranslational Translocation in Bacteria. Mol. Cell, 41, 343-353.
54. Wilson, D.N. and Beckmann, R. (2011) The Ribosomal Tunnel as a Functional Environment for Nascent Polypeptide Folding and Translational Stalling. Curr. Opin. Struct. Biol., 21, 274-282.
A) Secondary domains of the 23S rRNA of T. thermophilus. The six secondary structural domains of the 23S rRNA are distinguished by color: Domain I in gray, Domain II in brown, DIII in pink (core) and blue (appendage), Domain IV in yellow, Domain V in purple, and Domain VI in orange. B) Tertiary interactions and phosphate–Mg2+–phosphate linkages within Domain III. Each first shell Mg2+–phosphate interaction is indicated by a yellow circle. The yellow lines between the circles are the phosphate–Mg2+–phosphate linkages. The Mg-µc involves nucleotides A1395, A1603 and C1604, bridging helices 51 and 53 and is highlighted in green.The black lines are rRNA-rRNA interactions as calculated by FR3D [38]. C) DIIIcore with the stem-loop sequence (highlighted in gray) used to connect the two fragments. SHAPE reactivity data for DIIIcore in 250 mM Na+ are indicated, where nucleotides highlighted by blue circles are unreactive.
A) Space-filling representation of DIII in three orientations, illustrating the subdomain architecture. DIIIcore is shown in magenta and DIIIH54-59 is shown in blue. B) DIII interacts with L23 (green), which is associated with DIIIcore but not DIIIH54-59. Coordinates were extracted from the LSU (PDB 2J01).
Effect of Mg2+ on the SHAPE reactivity of DIIIcore (blue) and DIII (brown). The vertical axis represents SHAPE reactivities and the horizontal axis represents nucleotide position as indicated in Figure 1. A) DIII and DIIIcore reactivity in 250 mM Na+. B) DIII and DIIIcore reactivity in 250 mM Na+ and 10 mM Mg2+.
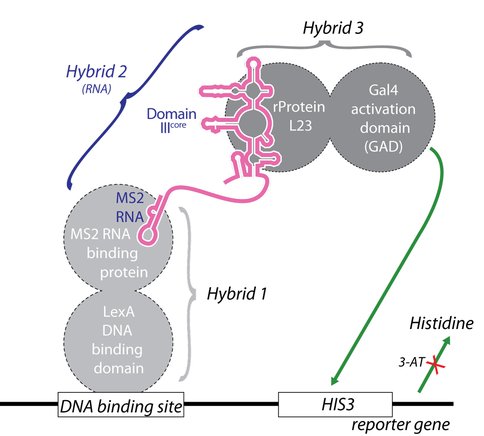
Schematic diagram of the yeast-three-hybrid assay for detecting and analyzing RNA-protein interactions. In yeast strain YBZ-1, the HIS3 reporter gene is under the control of the LexA operator. Hybrid 1, a LexA/MS2 coat protein fusion, binds to the LexA operator. The MS2 coat protein domain binds tightly to the MS2 sequence of the hybrid RNA, which contains the MS2 RNA and RNA sequence of interest, e.g., DIII or DIIIcore. In Hybrid 3, the protein of interest (e.g., L23) is fused to the yeast GAL4 transcriptional activation domain (GAD). In vivo RNA-protein binding completes Hybrid 2, resulting in expression of the HIS3 reporter gene. The strength of RNA-protein binding is determined by resistance to 3-Amino-1,2,4-triazole (3-AT), a competitive inhibitor of the HIS3 product, imidazoleglycerol-phosphate dehydratase (Stumpf et al, 2008).
Yeast three-hybrid assay results for 3-AT resistance of yeast strain YBZ-1 expressing Domain III-MS2 or DIIIcore -MS2 and GAD-L23, as well as positive control p50/p53. Cell growth (measured at O.D. 630 nm after 48 hr) is plotted against increasing 3-AT concentration. Negative controls are MS2 RNA alone (lacking DIII or DIIIcore) assayed with L23 and p53. Activity from negative controls is background signal.
Continuous variation of fluorescence intensity versus the mole fraction (X) of L23peptide. A) The second component is DIII rRNA. B) The second component is DIIIcore rRNA. The trend lines intersect at x=0.53 for DIII and at x=0.55 for DIIIcore, consistent with a 1:1 stoichiometry of interaction of L23peptide with each RNA.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Shreyas Athavale
Postdoc
Dana Cook-Schneider
Postdoc
Chiaolong Hsiao
Postdoc
Jessica Bowman
Research Staff
Eli Hershkovits
Research Staff
Eric O'Neill
Research Staff
Anton Petrov
Research Staff
Chad Bernier
Graduate Student
Jared Gossett
Graduate Student
Poorna Roy
Graduate Student
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules
Objective 4.1
Earth's early biosphere.
Objective 4.2
Production of complex life.