2012 Annual Science Report
 Georgia Institute of Technology
Reporting | SEP 2011 – AUG 2012
Georgia Institute of Technology
Reporting | SEP 2011 – AUG 2012
An Atomic Level Description of the Specific Interactions Between Nascent Peptide and Ribosome Exit Tunnel
Project Summary
The ribosome exit tunnel is an ancient path that must be traveled by all peptides/proteins synthesized by the ribosome. We have synthesized peptolides and demonstrated their potential as probes to decipher the interaction between the nascent peptide and the exit tunnel. This study has furnished vital information about the path of travel of peptides attached to the flag-pole moiety of a ketolide. In continuation of our study, we have designed and commenced the synthesis of a series of oxazolidinone-peptide conjugates (Zyvotides) that places nascent peptide at a different window to the exit tunnel. We will characterize the interaction of these zyvotides with the ribosome exit tunnel using the tools we have developed during our investigation of the peptolides.
Project Progress
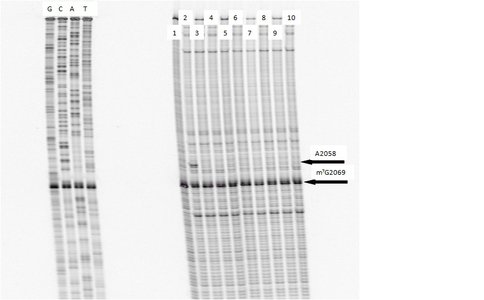
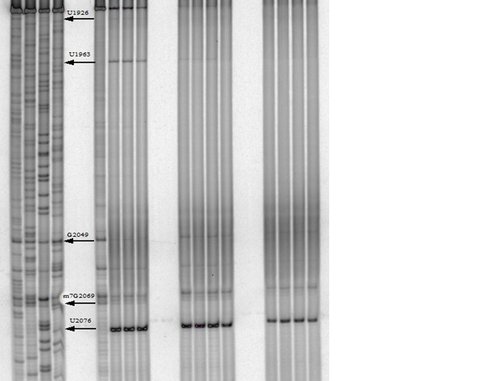
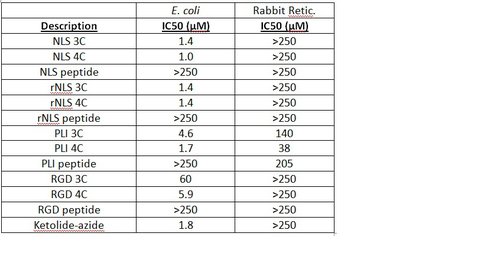
One of our primary interests is to elucidate the nature of the interaction between the ribosome peptide exit tunnel and the nascent peptide before breeching the extraribosomal interface. In doing so, we will be able to gain a better understanding of the molecular basis for the choice of nucleic acid as exit tunnel construction materials. Using molecular probes – peptolides – that we have developed, we have characterized the interaction between varied peptide sequences and key residues within the exit tunnel and the space adjoining the entrance to the exit tunnel and the peptidyl transferase site. This was accomplished by cell-free translation inhibition assays (Table 1), footprinting experiments of the ribosomal RNA (rRNA) through dimethyl sulfate (DMS) and 1-cyclohexyl-(2-morpholino-ethyl)carbodiimide metho-p-toluene sulfonate (CMCT) modification in the presence of the peptolides (Figs. 1 & 2). Our footprinting data strongly suggest that the peptide moieties of all the peptolide studied preferred a path of travel where they are oriented back to the peptidyl transferase site. This observation is consistent with crystallographic data on telithromycin (ketek), the ketolide template for all our peptolides, showing that it could adopt a conformation which has its flag-pole moiety oriented toward the peptidyl transferase site.1-3
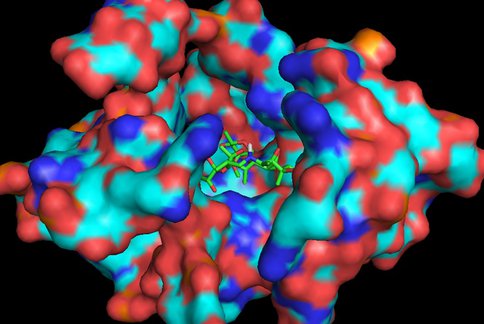
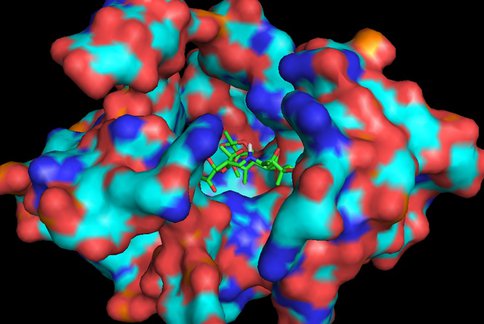
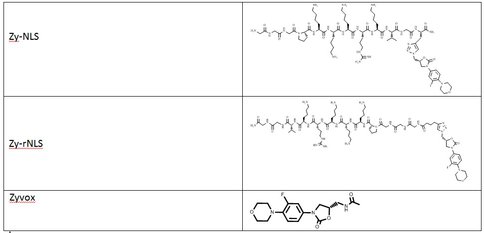
In continuation of our study, we have designed and commenced the synthesis of a series of oxazolidinone-peptide conjugates (Zyvotides) (Table 2) that places nascent peptide at a different window to the exit tunnel, thereby affording a path to the tunnel that is quite different from the peptolides’. We will characterize the interaction of these Zyvotides with the ribosome exit tunnel using the tools we have developed during our investigation of the peptolides. Additionally, we are designing another generation of peptolides to address the deficiency of the 1st generation pepolides which were built off of telithromycin flag-pole moiety. We had anticipated that the flexibility of the flag-pole moiety would afford an easy placement of the peptides down the exit tunnel, mimicking a growing peptide. As stated earlier, it seems flipping the peptide back to the peptidyl transferase site is preferred. By replacing the telithromycin scaffold with that of azithromycin, an azalide, the flexibility of the flag-pole moiety is removed. Crystal structures of both H. marismortui and T. thermophilus with azithromycin bound show that the proposed site of peptide attachment, N10, has an expansive space that would easily permit modification (Figs. 3-6). The loss of the flexibility afforded by telithromycin will allow the peptide probe to be guided down the exit tunnel.
References
1) Berisio, R.; Harms, J.; Schluenzen, F.; Zarivach, R.; Hansen, H. A.; Fucini, P.; Yonath, A. J. Bacteriol. 2003, 185, 4276-4279.
2) Tu, D.; Blaha, G.; Moore, P. B.; Steitz, T. A. Cell 2005, 121, 257–270.
3) Dunkle, J. A.; Xiong, L.; Mankin, A. S.; Cate, J. H. D. PNAS, 2010, 107, 17152-17157.
DMS Footprinting of 23S rRNA with primer 2180. Dideoxy sequencing lanes G,C,A and T followed by lanes: 1) unmodified rRNA, 2) DMS modified rRNA, Lanes 3-10) rRNA DMS modified in the presence of Clarithromycin, ketolide-azide, 3C NLS, 4C NLS, 3C rNLS, 4C rNLS, 3C PLI, and 4C PLI respectively.
CMCT Footprinting of 23S rRNA with primer 2180. Dideoxy sequencing lanes G,C,A and T followed by lanes: 1) unmodified rRNA, 2) CMCT modified rRNA, Lanes 3-10) rRNA CMCT modified in the presence of Clarithromycin, ketolide-azide, 3C NLS, 4C NLS, 3C rNLS, 4C rNLS, 3C PLI, and 4C PLI respectively.
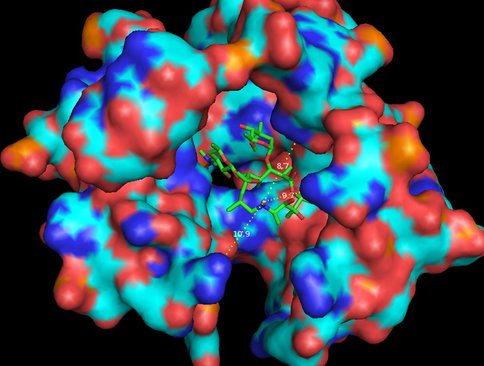
Haloarcula marismortui with azithromycin bound (1M1K). 12Å bubble around azithromycin looking back to PTC. Proposed site of modification in white.
Haloarcula marismortui with azithromycin bound (1M1K). 12Å bubble around azithromycin looking back to PTC. Proposed site of modification in white.
Same as Figure 3 rotated forward 90O showing measurements to nearest residues.
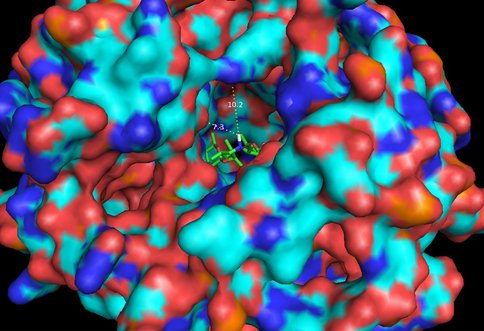
Thermus thermophilus with azithromycin bound (3OHZ). 12Å bubble around azithromycin looking back to PTC. Proposed site of modification in white.
Same as Figure 5 expanded to 20Å showing measurements to nearest residues.
IC50 values of in vitro, cell free assays performed on peptolide compounds.
Zyvotides synthesized for testing
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Subhasish Tapadar
Postdoc
Josh Canzoneri
Graduate Student
Arren Washington
Graduate Student
Eric Raftery
Undergraduate Student
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules