2010 Annual Science Report
 VPL at University of Washington
Reporting | SEP 2009 – AUG 2010
VPL at University of Washington
Reporting | SEP 2009 – AUG 2010
Postdoctoral Fellow Report: Steven Mielke
Project Summary
This project seeks to resolve the long-wavelength limit of oxygenic photosynthesis in order to constrain the range of extrasolar environments in which spectral signatures of biogenic oxygen might be found, and thereby guide future planet detecting and characterizing observatories.
Project Progress
Investigations forming the basis of my NAI/NPP fellowship aim to resolve the long-wavelength limit of oxygenic photosynthesis to advance the goals of the VPL task described in detail in the “Thermodynamic Efficiency of Electron-Transfer Reactions in the Chlorophyll d-Containing Cyanobacterium, Acaryochloris marina“ project report. To this end, we are engaged in experimental and theoretical studies of photochemistry in the cyanobacterium, Acaryochloris marina, which uses an unusual chlorophyll—chlorophyll (Chl) d rather than Chl a—to perform oxygenic photosynthesis in far-red light environments [1].
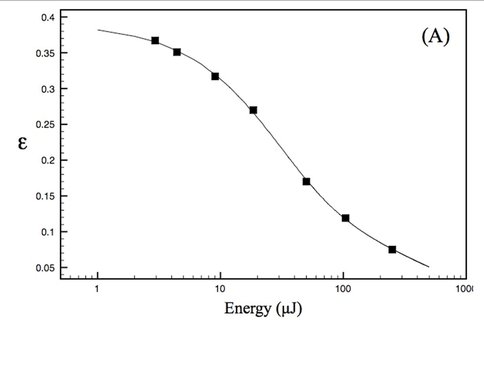
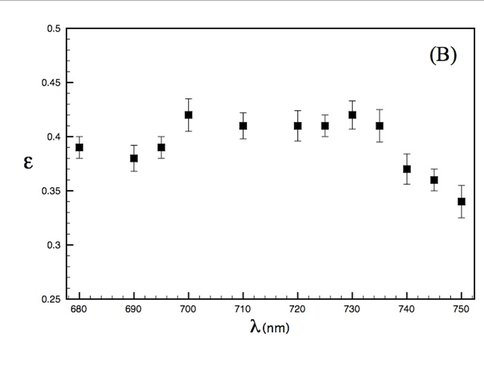
Our experimental studies, carried out in collaboration with Prof. David Mauzerall, Rockefeller University, and supported by an NAI DDF grant, seek to elucidate thermodynamics of photochemistry in A. marina, using pulsed, time-resolved photoacoustics (PTRPA), which provide simple, fast, and accurate measurements of thermodynamic quantities associated with photosynthetic reactions on ns–ms timescales [2]. Specifically, we have (Years 1 and 2) obtained the millisecond-timescale thermal efficiency of photosynthesis in A. marina intact cells over a range of wavelengths from 670 to 800 nm, where thermal efficiency, η, is defined as the ratio of energy stored in chemical products to photon energy absorbed. Representative results are shown in Figure 1. Our principal conclusion to date (results currently being prepared for publication) is that the efficiency of photosynthesis in A. marina at 710 nm, the wavelength of peak absorbance by our samples-is comparable to that in oxygenic species that employ Chl a and inhabit typical light environments. This indicates that photochemistry in Acaryochloris is not limited by the organism’s use of far-red photons, in turn indicating that wavelengths fundamentally limiting to oxygenic photosynthesis are longer than those utilized by A. marina, extending the range of possible extrasolar environments for biogenic oxygen. Investigations during Year 3 will quantify A. marina’s μs-timescale thermal efficiency, and photosystem-specific contributions to energy storage, as a function of wavelength, λ, and interpret the far-red drop in efficiency for λ > 735 nm, leading to at least one additional publication.
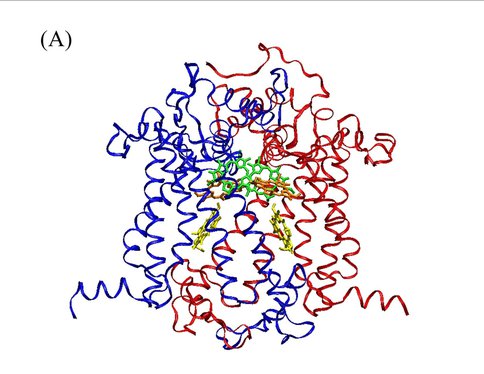
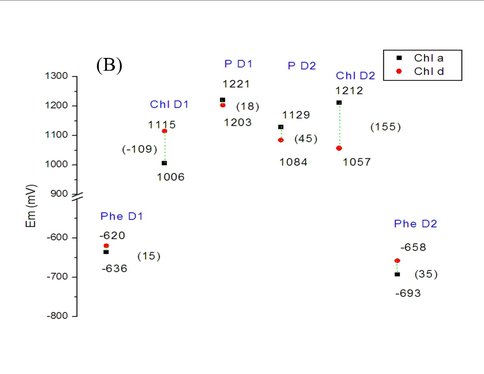
Our theoretical studies, carried out in collaboration with Prof. Marilyn Gunner, City College of New York, and supported by a DOE Energy Biosciences grant (Prof. Gunner, PI), seek to elucidate redox properties of photochemistry in A. marina, using multi-conformation continuum electrostatics (MCCE) methods. MCCE models protein electrochemistry by Monte Carlo sampling side-chain conformations to obtain charge states, and thereby, for example, midpoint redox potentials (Ems) of ionizable residues and ligands [3]. Specifically, we have (Years 1 and 2): (1) using known A. marina protein sequences, developed a homology model of the organism’s photosystem (PS) II reaction center (D1 and D2 subunits), inclusive of the six core cofactors (see Figure 2A); (2) in both the A. marina model structure and a known Thermosynechococcus elongatus PSII crystal structure (PDB ID 3bz2), calculated Ems of these cofactors using MCCE, where, in both structures, chlorophylls were designated alternately as Chl a or Chl d; and (3) compared the geometries of amino acid side-chains near the functional group that is modified in Chl d, relative to Chl a, in both the A. marina and T. elongatus structures. Our results to date, currently being prepared for publication, indicate that for none of the cofactors, in either structure, does replacing Chl a with Chl d produce a significant shift in Em. The largest differences are seen for the accessory chlorophylls, where the presence of Chl d shifts the Ems by ~100–150 mV (see Figure 2B). Our results further indicate that there are no significant differences in hydrogen-bonding propensity owing to local side-chain identities or geometries (data not shown). Taken together, these results suggest that the switch from Chl a to d affects the excited state, but not the cation product, of the primary electron donor. If confirmed, this implies that A. marina has not needed to adjust acceptor-side redox chemistry significantly to carry out efficient water oxidation in far-red light environments, supporting the conclusion of our experimental study. Investigations during Year 3, leading to at least one additional publication, will include further study of binding-site properties that might provide insight into the mechanism of A. marina’s selection of Chl d over Chl a; modeling of electron-transfer kinetics to complement both theoretical and experimental investigations of thermodynamics; and initiating a comprehensive study of PSI in Acaryochloris analogous to that of PSII.
(A) Saturation profile of A. marina whole cells obtained by PTRPA at a pulse wavelength of 730 nm. The thermal efficiency, , decreases with increasing flux because of saturation of the sample obeying the cumulative one-hit Poisson function. The curve is a nonlinear least-squares fit of this function to the PTRPA data (2 0.001), indicating a total (x-intercept) efficiency of ~39% for this data set.
Thermal efficiency in A. marina whole cells as a function of pulse wavelength between 680 and 750 nm (resolution 2 nm). For > 735 nm, steadily decreases as photon energies fall below those of the excited-state reaction center traps.
(A) Homology model of D1 (blue) and D2 (red) protein subunits of PSII in A. marina. The model includes the primary and accessory chlorophylls (green and orange, respectively), and the primary electron acceptor, pheophytin (yellow). Protein sequence identity with other cyanobacteria, e.g., T. elongatus, is high (~85%), leading to models with high similarity to known structures.
(B) Midpoint redox potentials of the six cofactors within the A. marina model structure, calculated from MCCE titrations. The largest shift in Em from replacing Chl a with Chl d (the divinyl group in the former with a formyl group) is seen for the accessory chlorophylls, ChlD1 and ChlD2, and is attributable primarily to differences in desolvation energy. Similar results were obtained for the cofactors within the T. elongatus crystal structure, 3bz2 (data not shown).
Publications
- Mielke, S., Dong, M., Kiang, N. & Gunner, M. (In Press). Midpoint redox potentials of PSII cofactors in the cyanobacterium, Acaryochloris marina, calculated by multi-conformation continuum electrostatics (MCCE).
- Mielke, S., Kiang, N., Blankenship, R. & Mauzerall, D. (2010). “Thermal Efficiency of Photosynthesis in the Cyanobacterium, Acaryochloris marina,” – Oral Presentation and Poster. Eastern Regional Photosynthesis Conference. Marine Biological Laboratory, Woods Hole, MA.
- Mielke, S., Kiang, N., Chen, M., Blankenship., R.E. & Mauzerall, D. (In Press). Thermal efficiency of photosynthesis in the cyanobacterium, Acaryochloris marina.
- Mielke, S., Kiang, N., Gunner, M., Blankenship, R. & Mauzerall, D. (2009). “Photosynthetic Electron-Transfer in the Cyanobacterium, Acaryochloris marina,” – Oral Presentation. NAI Executive Council Meeting. Yellowstone National Park, Wyoming.
- Mielke, S.K.N., Blankenship, R., Gunner, M. & Mauzerall, D. (2010). “Photosynthetic Electron-Transfer in the Cyanobacterium, Acaryochloris marina,” – Poster and Lightning Talk. Astrobiology Science Conference 2010. League City, Texas.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Robert Blankenship
Co-Investigator
Marilyn Gunner
Co-Investigator
Nancy Kiang
Co-Investigator
David Mauzerall
Co-Investigator
-
RELATED OBJECTIVES:
Objective 5.1
Environment-dependent, molecular evolution in microorganisms
Objective 6.1
Effects of environmental changes on microbial ecosystems
Objective 6.2
Adaptation and evolution of life beyond Earth
Objective 7.2
Biosignatures to be sought in nearby planetary systems