2010 Annual Science Report
 Montana State University
Reporting | SEP 2009 – AUG 2010
Montana State University
Reporting | SEP 2009 – AUG 2010
Molecular Paleontology of Iron-Sulfur Enzymes
Project Summary
In this project we are attempting to trace back in the evolutionary record using specific genetic events as markers. We are using specific gene fusion and gene duplication events in the genetic record to place a chronological sequence to the advent of nitrogen fixation, certain modes of hydrogen metabolism, and both anoxygenic and oxygenic photosynthesis.
Project Progress
We have been using a variety of different approaches to examine the evolutionary trajectories and evolutionary origins of nitrogen fixation, hydrogen metabolism, and photosynthesis. These are all three complicated biochemical processes that instead of being mediated by simply one or two gene product(s) require large suites of genes. For these three processes the main biochemical machinery; nitrogenase for nitrogen fixation, hydrogenase for hydrogen metabolisms, photosystems for photsynthesis, require specific gene products for the synthesis and assembly of complex prosthetic groups that are integral and essential for their respective functions. The requirement for numerous gene products for these enzymes allows use to probe the phylogenetic trajectories of suite of genes to trace evolutionary relatedness and trajectories and is a much more robust approach than simply basing inheritance on a single gene. In addition, the processes of nitrogen and photosynthesis are intimately linked since the nitrogenase structural genes and key enzymes in dark chlorophyll biosynthesis are related by gene duplication. For hydrogen metabolism [FeFe]-hydrogenases and [NiFe]-hydrogenases are related to our strongly related to other enzymes. The [FeFe]-hydrogenase are related to a protein called Nar1p that is only found in eukaryotes and implicated in being involved in iron-sulfur cluster biosynthesis in the cytosol. The [NiFe]-hydrogenases are related to respiratory complex I suggesting that H2 itself may have been a predecessor to NADH in early metabolism. Probing evolutionary trajectories of the processes that are central to the ABRCs thrust is rapidly developing has been thus far quite fruitful. Using this approach we have been able to implicate that the complex biosynthetic machinery involved in the synthesis of the active site complex iron-sulfur clusters and maturation of active Mo-nitrogenase and [FeFe]-hydrogenase was adapted after the structural genes were apparently recruited for the process. Further the results indicate, as perhaps anticipated in retrospect, that these enzymes as we see them today with their complicated cofactors are not likely to be that old. We have also to infer in preliminary work that the active sites of [NiFe]-hydrogenases and carbon monoxide dehydrogenases probably follow a distinctly different evolutionary path and are likely to be more ancient. For Mo-nitrogenase we are able to infer that the most ancient of extant forms of this active enzyme occur in the methanogenic Archaea. The results have paved the way for challenging paradigms regarding the evolutionary path of complex iron-sulfur enzymes and have allowed us to rationalize a clear hypothetical path that we can examine through further experimentation and analysis.
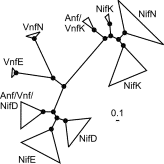
Phylogenetic tree of Nif/Vnf/AnfD, K, E, and N deduced amino acid sequences. The width and depth of clades proportionally reflect the number and the diversity of sequences in that clade. Bar equals 1 substitution per 10 sequence positions. Black circles denote 90–100% posterior probability (PP) at a given node. The results highlight the relationship between the nitrogenase structural genes NifDK and the evolutionarily related biosynthetic gene products NifDK. The results provide compelling proof that the the biosynthetic gene product E (nest in NifD sequences in the phylogram) was derived from NifD by duplication and that NifN (nest in NifK sequences in the phylogram) was derived from NifK. These results indicate that these elements (NifEN) of the biosynthetic apparatus for the nitrogenase iron-molybdenum cofactor emerged evolutionary after the NifDK structural genes. These results also represent the preliminary basis for suggested Mo-nitrogenase as we know it today arose in methanogens.
Unrooted radial phylogram is inset. ME denotes mitochondriate eukaryotes, AE denotes amitochondriate eukaryotes, and no designation denotes bacterial sequences. The designations and numerals following designations correspond in both figures. Closed circles denote 90-100% bootstrap support (BS); gray circles denote 80-89% BS; open circles denote 70- 79% BS support; no symbol indicates <69% BS. The results indicate that [FeFe]-hydrogenases are likely to have an ancestor that lack the complicated iron-sulfur cluster prosthetic group and that the gene products involved in cluster biosynthesis were likely recruited at the time or after the structural was recruited for hydrogen metabolism.
Publications
-
Boyd, E. S., Anbar, A. D., Miller, S., Hamilton, T. L., Lavin, M., & Peters, J. W. (2011). A late methanogen origin for molybdenum-dependent nitrogenase. Geobiology, 9(3), 221–232. doi:10.1111/j.1472-4669.2011.00278.x
-
McGlynn, S. E., Boyd, E. S., Shepard, E. M., Lange, R. K., Gerlach, R., Broderick, J. B., & Peters, J. W. (2009). Identification and Characterization of a Novel Member of the Radical AdoMet Enzyme Superfamily and Implications for the Biosynthesis of the Hmd Hydrogenase Active Site Cofactor. Journal of Bacteriology, 192(2), 595–598. doi:10.1128/jb.01125-09
-
Mulder, D. W., Boyd, E. S., Sarma, R., Lange, R. K., Endrizzi, J. A., Broderick, J. B., & Peters, J. W. (2010). Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydAΔEFG. Nature, 465(7295), 248–251. doi:10.1038/nature08993
-
Shepard, E. M., Boyd, E. S., Broderick, J. B., & Peters, J. W. (2011). Biosynthesis of complex iron–sulfur enzymes. Current Opinion in Chemical Biology, 15(2), 319–327. doi:10.1016/j.cbpa.2011.02.012
-
Soboh, B., Boyd, E. S., Zhao, D., Peters, J. W., & Rubio, L. M. (2010). Substrate specificity and evolutionary implications of a NifDK enzyme carrying NifB-co at its active site. FEBS Letters, 584(8), 1487–1492. doi:10.1016/j.febslet.2010.02.064
- Glass, J.B., Boyd, E.S., Romaniello, S.J. & Anbar, A.D. (2010). Evolutionary metallomics: a nickel for nitrogenase. In review.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
John Peters
Project Investigator
Joan Broderick
Co-Investigator
Eric Boyd
Postdoc
Trinity Hamilton
Doctoral Student
David Mulder
Doctoral Student
Rachel Lange
Undergraduate Student
-
RELATED OBJECTIVES:
Objective 3.1
Sources of prebiotic materials and catalysts
Objective 3.2
Origins and evolution of functional biomolecules
Objective 3.3
Origins of energy transduction
Objective 3.4
Origins of cellularity and protobiological systems
Objective 4.1
Earth's early biosphere.
Objective 5.1
Environment-dependent, molecular evolution in microorganisms
Objective 5.2
Co-evolution of microbial communities
Objective 5.3
Biochemical adaptation to extreme environments
Objective 6.1
Effects of environmental changes on microbial ecosystems
![Phylogenetic Relationships Between HydA ([FeFe]-hydrogenases and Nar1p](../../../../media/site-content/reports/2010/mon/rzppekdrmk_qgerddtjuq_nature.jpg.484x0_q85.jpg)