2010 Annual Science Report
 Montana State University
Reporting | SEP 2009 – AUG 2010
Montana State University
Reporting | SEP 2009 – AUG 2010
Complex Chemical Networks in Astrobiology
Project Summary
Many of the building blocks for prebiotic chemistry form in dark molecular clouds — dense regions of interstellar gas and dust from which stars and planetary systems are also born. Sophisticated chemical models have been developed to understand the complex network of reactions that can convert simple precursor molecules to the complex organics that are often found in meteorites and comets. We have begun to apply the tools of network theory — a branch of mathematics that studies interconnected complex systems from cellular metabolism to Facebook — to gain more insight into the structure of these interstellar chemical networks. This approach allows us to make comparisons between complex chemical networks in biology, astronomy, and planetary science, searching for unifying general principles and critical differences between living and non-living systems.
Project Progress
A recurring theme in astrobiology is the emergence of chemical complexity from systems containing a large number of species and reactions. The environments involved are greatly varied – including the interstellar medium, planetary atmospheres, solution-phase abiotic environments and living cells – but these diverse systems may also include shared features. An auxiliary project within the ABRC has involved the computational exploration of the architecture of these complex chemical networks. Thus far, our primary test case has been the chemical reaction network of dark molecular clouds, as represented in the UMIST Database for Astrochemistry (UDfA). This has required abstracting the details of the chemical system to a graph representation, in which species (and sometimes reactions) are represented by nodes with edges representing the relationships between them. These methods have been developed, to a large degree, within the bioinformatics community and have been applied to the analysis of metabolic networks. We have found that the astrochemical reaction network shows a large-scale structure rather different from that seen in biochemical networks – possibly attributable to the fact that they operate in a near-equilibrium regime rather than the far-from-equilibrium dynamics typical in biology – and that several features of the network structure shift significantly during the course of cloud evolution. The application of network-theoretical tools to non-biological chemical networks has not been extensively explored in the past and may provide a fruitful venue for comparing the properties of living and non-living chemical systems. Current efforts in this direction include the development of theoretical tools for the analysis of directed bipartite networks containing both chemical species and reactions; these tools have shown interesting preliminary results with the UDfA reaction system. Within the next year, we hope to apply these same tools to the metabolic network of E. coli, as well as the atmospheres of Earth and Titan.
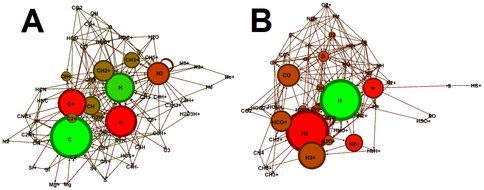
(A) Graph generated from the cloud chemistry simulation at 10^2 y. Nodes correspond to chemical species present in the simulation, while an edge between nodes indicates that a reaction consumes one species and produces another. The size of nodes is proportional to their weighted degree, and they are color-coded such that rapidly-produced species are green while rapidly-destroyed species are red. Only the 5% most heavily-weighted edges and their corresponding nodes are shown. (B) Graph generated from the simulation at 10^7 y. Note that highly-reactive organic cations (CH+, CH2+, CH3+, etc.) are prominent early in cloud evolution while more stable oxygen-bearing species (CO, O2, HCO+, etc.) appear at later stages. Network diagrams such as these provide a simple visual representation of the relationships between species in a complex reaction network.
Publications
-
Jolley, C. C., & Douglas, T. (2010). A NETWORK-THEORETICAL APPROACH TO UNDERSTANDING INTERSTELLAR CHEMISTRY. The Astrophysical Journal, 722(2), 1921–1931. doi:10.1088/0004-637x/722/2/1921
-
Jolley, C. C., & Douglas, T. (2010). Ion Accumulation in a Protein Nanocage: Finding Noisy Temporal Sequences Using a Genetic Algorithm. Biophysical Journal, 99(10), 3385–3393. doi:10.1016/j.bpj.2010.09.001
-
Jolley, C. C., Uchida, M., Reichhardt, C., Harrington, R., Kang, S., Klem, M. T., … Douglas, T. (2010). Size and Crystallinity in Protein-Templated Inorganic Nanoparticles. Chem. Mater., 22(16), 4612–4618. doi:10.1021/cm100657w
-
Jolley, C., Klem, M., Harrington, R., Parise, J., & Douglas, T. (2011). Structure and photoelectrochemistry of a virus capsid–TiO 2 nanocomposite. Nanoscale, 3(3), 1004–1007. doi:10.1039/c0nr00378f
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Trevor Douglas
Co-Investigator
-
RELATED OBJECTIVES:
Objective 3.1
Sources of prebiotic materials and catalysts
Objective 3.2
Origins and evolution of functional biomolecules