2010 Annual Science Report
 Massachusetts Institute of Technology
Reporting | SEP 2009 – AUG 2010
Massachusetts Institute of Technology
Reporting | SEP 2009 – AUG 2010
Origins of Multicellularity
Project Summary
By comparing animal genomes with genomes from their closest living relatives, the choanoflagellates, we can reconstruct the genome composition of the last common ancestor of animals.
Project Progress
To gain insight into the origin of animal multicellularity, we aim to reconstruct the genome of the last common ancestor of animals and their sister group, the choanoflagellates. We have previously led the sequencing of genomes from two choanoflagellates (Monosiga brevicollis (1) and Salpingoeca rosetta [www.broadinstitute.org/annotation/genome/multicellularity_project/Info.html]), both of which lie within one of the three major choanoflagellate clades (Figure 1) (2). In addition, the first sequence of a sponge was reported in the last year, providing an important point of comparison from an early-branching animal lineage (3). Nonetheless, these genomes are likely to provide an incomplete picture of the genome in the last common ancestor of animals and choanoflagellates [e.g. see (4)]. Therefore, we are currently sequencing the genomes and transcriptomes of representative choanoflagellates (Salpingoeca napiformis and Diaphanoeca grandis) from the two remaining unsampled choanoflagellate clades. In addition, we have sequenced the genome of a homoscleromorph sponge (Oscarella carmela), representing a sponge lineage that contains diagnostic features of eumetazoan biology.
The development of Illumina short read sequencing technology has allowed the production of draft genome assemblies from non-model organisms for relatively low cost. Starting with a single sponge larva we amplified and sequenced the ~54Mbp genome of the sponge O. carmela at more than 500x depth of coverage. The limitation of this approach is that the relatively small insert size of the sequenced library (400bp) precludes the assembly of scaffolds that contain low complexity or highly repetitive sequence motifs (e.g., intergenic regions and large introns). Thus, our final assembly was useful for gene discovery, but not for analyzing genome structure or non-coding DNA. Furthermore, we view the resultant gene set as a minimal estimate, not comprehensive due to the fact that some genes may not have assembled. Nonetheless, we found that >90% of previously sequenced Sanger ESTs were represented.
Choanoflagellates provide unique challenges for genome projects because they are co-cultured with prey bacteria, whose genomes can interfere with robust assembly of the target choanoflagellate genome. Despite this, we have been able to produce pilot genome assemblies for both S. napiformis and D. grandis in the past year. In addition, we will improve these assemblies over the coming year via additional genomic DNA and transcriptome sequencing.
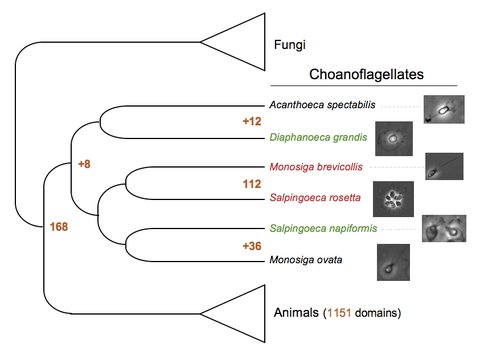
The current assemblies of the S. napiformis and D. grandis genomes provide useful datasets from which to expand our understanding of the biology of the last common ancestor of animals and choanoflagellates. To this end, we have initially focused on the identification of protein domains that are shared exclusively among choanoflagellates and animals. Protein domains have been successfully studied in choanoflagellates in our previous work, as they represent a unit of biological function and can be identified across long evolutionary distances (1). We have drawn three major conclusions from identifying protein domains in pilot assemblies of S. napiformis and D. grandis. First, we have discovered 56 protein domains in these two choanoflagellates that were previously thought to be specific to animals, thus extending their evolutionary history to the premetazoan era. Second, we have increased the number of protein domains inferred to be present in the common ancestor of choanoflagellates and animals by 50%, from the 112 domains previously found in M. brevicollis and S. rosetta to 168 domains. Third, many of these newly discovered protein domains represent key animal development and signaling pathways not previously observed outside of animals, such as the RHD domain, which is found exclusively in members of the immunologically important NF-κB family of transcription factors in animals. These findings speak to the importance of gene co-option and exon shuffling during the origin and evolution of animal signaling pathways.
As a complement to the global genome-sequencing approach, we are performing a more focused study on the ancestry of the hedgehog signaling pathway. The hedgehog protein, a key regulator of developmental patterning in bilaterian animals, signals through the primary cilium, which is equivalent to the choanoflagellate flagellum (5, 6). We have identified core components of the hedgehog signaling pathway in M. brevicollis that were previously known only from animals (1, 7, 8). Although a direct homolog of the hedgehog signaling ligand is absent in choanoflagellate, we have identified a membrane-integral cadherin protein (hedgling/MBCDH11) that has structural homology to the hedgehog signal domain (7). In the past year we have generated antibodies against MBCDH11 to test two discrete hypotheses: 1) that the N-terminal region with homology to the hedgehog signaling ligand is cleaved and functions as a diffusible signal, and; 2) that the membrane-proximal portion of the protein localizes to the apical flagellum — a pattern that would be consistent with an ancient signaling role for both the hedgling protein and the primary cilium. In addition, we have affinity purified fractions that specifically recognize the injected antigen, and have made significant strides towards validating that these antibodies are specific to hedgling in vivo; this is a unique challenge due to its exceptionally large size (~520KDa). M. brevicollis provides a simple system in which to study the ancestral function of hedgehog signaling and promises to yield insight into the role of this important developmental signaling pathway during the transition to multicellularity.
In addition to initial findings from M. brevicollis, we have recently made important insights into the ancestral complexity and assembly of the Hedgehog signaling pathway from sequencing the genome of S. rosetta and O. carmela. Specifically, we have learned that S. rosetta encodes another cadherin (for which there is an ortholog in M. brevicollis) that has a conserved, N-terminal Hh signal domain. This protein is significantly smaller and may provide a more accessible experimental target. Furthermore, we have discovered that O. carmela (unlike the genome of the sponge Amphimedon queenslandica) potentially encodes a bona-fide hedgehog ligand, in addition to a conserved hedling homolog. Consistent with this is the discovery of other pathway components present in O. carmela and absent in A. queenslandica and choanoflagellates, such as the protein dispatched that is required for ligand export. Together these data begin to paint a more complex portrait of the evolutionary history of the hedgehog signaling pathway than initially assumed. As these comparative genomic data become grounded in experimental research we stand to make unprecedented insights into the sequence by which this developmental signaling pathway was assembled and how its function evolved in concert with metazoan body plan diversity.
Choanoflagellates are the closest living relatives of animals, and protein domains shared exclusively between animals and choanoflagellates are inferred to be present in their common ancestor. Monosiga brevicollis and Salpingoeca rosetta (red) are choanoflagellates with previously sequenced genomes, and they both lie within one of the three major choanoflagellate clades. As part of this project, we selected Diaphanoeca grandis and Salpingoeca napiformis (green) for genome sequencing. Of the 1,151 domains previously thought to be specific to animals, 112 are found in either M. brevicollis or S. rosetta. This study increased that number by 36 protein domains found in the clade containing S. napiformis, 12 domains in D. grandis, and 8 domains found in both clades, for a total of 168 of the 1,151 previously animal-specific domains that we infer played a role in the biology of the common unicellular ancestor of choanoflagellates and animals. Phylogenetic tree adapted from (2).
1. N. King et al., Nature 451, 783 (Feb 14, 2008).
2. M. Carr, B. S. Leadbeater, R. Hassan, M. Nelson, S. L. Baldauf, Proc Natl Acad Sci U S A 105, 16641 (Oct 28, 2008).
3. M. Srivastava et al., Nature 466, 720 (AUG 5, 2010).
4. A. Sebe-Pedros, A. J. Roger, F. B. Lang, N. King, I. Ruiz-Trillo, Proc Natl Acad Sci U S A 107, 10142 (Jun 1, 2010).
5. T. Caspary, C. E. Larkins, K. V. Anderson, Dev Cell 12, 767 (May, 2007).
6. V. Singla, J. F. Reiter, Science 313, 629 (Aug 4, 2006).
7. M. Abedin, N. King, Science 319, 946 (Feb 15, 2008).
8. M. Adamska et al., Curr Biol 17, R836 (Oct 9, 2007).
Publications
-
Abedin, M., & King, N. (2010). Diverse evolutionary paths to cell adhesion. Trends in Cell Biology, 20(12), 734–742. doi:10.1016/j.tcb.2010.08.002
-
Sebe-Pedros, A., Roger, A. J., Lang, F. B., King, N., & Ruiz-Trillo, I. (2010). Ancient origin of the integrin-mediated adhesion and signaling machinery. Proceedings of the National Academy of Sciences, 107(22), 10142–10147. doi:10.1073/pnas.1002257107
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Scott Nichols
Postdoc
Daniel Richter
Graduate Student
-
RELATED OBJECTIVES:
Objective 4.2
Production of complex life.