2010 Annual Science Report
 Massachusetts Institute of Technology
Reporting | SEP 2009 – AUG 2010
Massachusetts Institute of Technology
Reporting | SEP 2009 – AUG 2010
Evolution and Development of Sensory and Nervous Systems in the Basal Branches of the Animal Tree
Project Summary
Animals interact with the world through complex sensory structures (eyes, ears, antennas, etc.), which are coordinated by collections of neurons. While the nervous and sensory systems of animals are incredibly diverse, a growing body of evidence suggests that many of these systems are controlled by similar sets of genes. We are looking at early branching and understudied lineages of the animal family tree (using the jellyfish Aurelia and the worm Neanthes respectively) to see if these animals use similar genes during neurosensory development as the better-studied fruit fly and mouse. This research is critical for determining which structures are shared between animals because of common ancestry (known as homologous structures) and those that evolved independently in different lineages. Ultimately, such research informs how morphologically and behaviorally complex animals evolve.
Project Progress
We continue our investigations into the early evolution of sensory organization in animals. Developmental gene expression in the jellyfish Aurelia is helping clarify the evolution of sense organs at the base of the animal tree. Work on the polychaete worm Neanthes is leading to a better understanding of the relationship between sensory structures and appendages in the early radiation of bilaterian animals.
During the 2009-2010 report period we have published research on the expression of Otx and POU classes of genes in the development of Aurelia sensory organization (Nakanishi et al 2010), and two papers on the neural organization and developmental gene expression, respectively, of Neanthes appendage development (Winchell et al a,b, 2010). Gene expression data suggests that sensory organ development in Aurelia is homologous to those of more complex bilaterian animals [figure 1]. Conversely, data from canonical “limb” genes in Neanthes do not support the homology of appendages across bilaterians [figure 2]. However, this work does suggest a relationship between the evolution of appendages and sensory structures. An additional book chapter reviewing the early evolution of sensory systems (Jacobs et al 2010), and a commentary in Science (Marshall and Jacobs 2009), were both published during the past year. Additional work on evolution in the coastal marine realm was also published.
The final projects of Nagayasu Nakanishi, a graduate student on the Advent of Complex Life project, have been published during this period as well as papers by Chris Winchell, who is slated to go on to a PostDoc at the Max Plank Institute in Tubingen, Germany. The first publication involving David Gold is now out and we expect David, and undergraduate students working with him, including Ignacio Navarette, to have a very successful year.
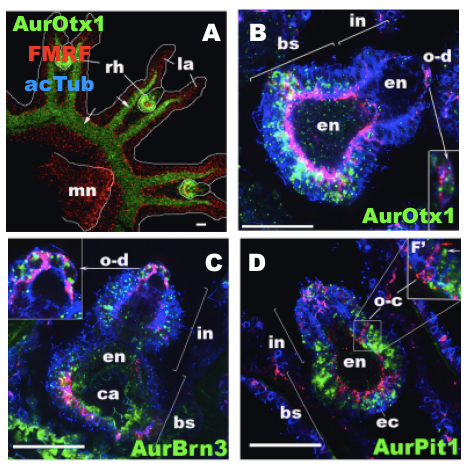
Figure 1: Expression of the genes Otx1, Pit1, and Brn3 in the jellyfish Aurelia sp. 1. These images focus on the rhopalia, which are the main sensory structures of the Aurelia jellyfish. Expression of the gene AurOtx1 [A, B] defines the major part of the oral neuroectodermal domain of the rhopalium, within which distinct populations of AurBrn3© and AurPit1 [D] expressing sensory cells develop. Thus, despite the unique attributes of rhopalial evolution, we suggest that the rhopalial nervous system of scyphozoan jellyfish involves similar patterns of gene expression that function in bilaterian cephalic structure and neuroendocrine system development. Images modified from Nakanishi et al. Evolution and Development; http://onlinelibrary.wiley.com/doi/10.1111/j.1525-142X.2010.00427.x/pdf
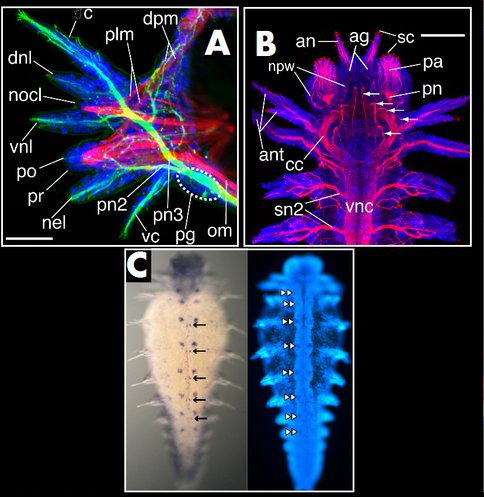
Figure 2: Nervous system and gene expression in the worm Neanthes. A: Third parapodium (limb) from a 15-segment Neanthes arenaceodentata juvenile. Neurons are labeled green with acetylated α-tubulin; muscle is labeled red with F-actin; cell nuclei are blue. From Winchell et al. Frontiers in Zoology 2010, 7:17; http://www.frontiersinzoology.com/content/7/1/17 B: 15-segment juvenile. Neurons are labeled with acetylated α-tubulin in red; cell nuclei are blue. From Winchell et al. Frontiers in Zoology 2010, 7:17; http://www.frontiersinzoology.com/content/7/1/17 C: Left panel shows apterous gene expression in a 10-segment juvenile (purple); right panel (same specimen) shows labeling of cell nuclei. apterous is expressed in ventral nerve cord ganglia in large lateral cell clusters (double arrowheads), and more posteriorly in smaller, medial cell clusters (arrows). Unpublished results.
Publications
-
Marshall, C. R., & Jacobs, D. K. (2009). Flourishing After the End-Permian Mass Extinction. Science, 325(5944), 1079–1080. doi:10.1126/science.1178325
-
Nakanishi, N., Yuan, D., Hartenstein, V., & Jacobs, D. K. (2010). Evolutionary origin of rhopalia: insights from cellular-level analyses of Otx and POU expression patterns in the developing rhopalial nervous system. Evolution & Development, 12(4), 404–415. doi:10.1111/j.1525-142×.2010.00427.x
-
Winchell, C. J., Valencia, J. E., & Jacobs, D. K. (2010). Confocal analysis of nervous system architecture in direct-developing juveniles of Neanthes arenaceodentata (Annelida, Nereididae). Frontiers in Zoology, 7(1), 17. doi:10.1186/1742-9994-7-17
-
Winchell, C. J., Valencia, J. E., & Jacobs, D. K. (2010). Expression of Distal-less, dachshund, and optomotor blind in Neanthes arenaceodentata (Annelida, Nereididae) does not support homology of appendage-forming mechanisms across the Bilateria. Dev Genes Evol, 220(9-10), 275–295. doi:10.1007/s00427-010-0346-0
- Jacobs, D.K., Gold, D.A., Nakanishi, N., Yuan, D., Camara, A., Nichols, S.A. & Hartenstein, V. (2010). Basal Metazoan Sensory Evolution. In: Desalle, B.S.a.R. (Eds.). Key Transitions in Animal Evolution. CRC Press.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Nicole King
Collaborator
Kevin Peterson
Collaborator
Ignacio Navarette
Undergraduate Student
Dhruv Patel
Undergraduate Student
Andrew Suh
Undergraduate Student
Jonathan Valencia
Undergraduate Student
-
RELATED OBJECTIVES:
Objective 4.1
Earth's early biosphere.
Objective 4.2
Production of complex life.