2010 Annual Science Report
 Georgia Institute of Technology
Reporting | SEP 2009 – AUG 2010
Georgia Institute of Technology
Reporting | SEP 2009 – AUG 2010
Ribosome Paleontology
Project Summary
We are inventing methodologies to determine the chronology of ribosomal origin and evolution. Our premise, which is generally accepted, is that substantial information relating to the origins and early development of the translation machinery remains imprinted in the ribosome: in the sequences, folding, assembly, molecular interactions, and functions of the ribosome’s various macromolecules and small molecule affectors. To this end, we are developing new methods for ribosomal paleontology. We are using these methods to determine the relative ages of ribosomal components and subsystems, and to understand fundamental aspects of the folding and assembly of RNA and protein. We will develop timelines for the history of the ribosome as a whole, as well as for various sub-processes such as initiation, termination, and translocation. The results of these studies will interface ribosomal history with other keys relating to the origin of life, including the origin of proteins and RNA, the emergence of the genetic code, the origin of chirality, and the nature of the last common ancestor.
Project Progress
It is now generally accepted that the peptidyl transferase center (PTC) and parts of the large subunit (LSU) assembled and commenced catalytic condensation of amino acids prior to association with the small subunit. As Fox described 1, 2, and recently discussed at ABSICON and the NAI Workshop Without Walls, this chronology has the important implication that the ancestral ribosome synthesized non-coded oligomers. The non-coded products are proposed to be peptide or peptide-like (racimates, mixed peptides, esters, thiolesters, etc). The ability to synthesize coded peptides or proteins was unlikely to be a driving force behind the emergence of the ribosome.
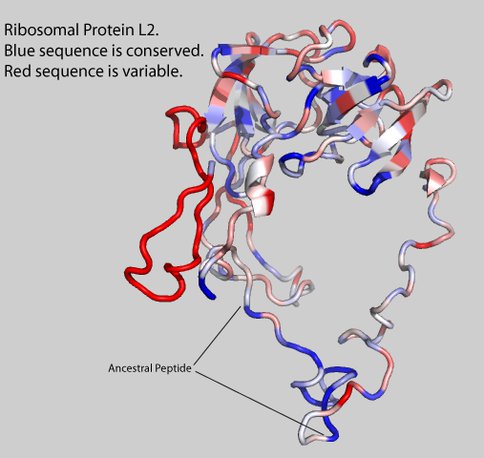
Over the past year Williams’ group developed the hypothesis that embedded in ribosomal protein L2 (rProtein L2, Figure 1) is biology’s oldest peptide. We hypothesize that this peptide is a molecular fossil of a non-coded product of the ancestral LSU. This peptide, or fragments of it, associated with the rRNA, conferring stability and protection from degradation. Using 130 species chosen to uniformly sample the phylogenetic tree, we investigated the alignment, consensus sequences, and the degree of conservation of sequence, conformation, and interactions. The consensus sequence of rProtein L2 is shown in Table 1. The proposed ancestral peptide is indicated.
Table 1a,b,c,d: Ribosomal protein L2 consensus sequence
MGKSLLEQRG GRNNSGRVTT RHRGGGHKRR YRIIDFKRNK DGIPGKVEDI EHDPNRSAPI
ALVHYEDGEK RYILAPEGLK VGDTIMCGPD APIKPGNALP LGNIPEGTIV HNIEMKPGDG
GQLARSAGTY AQIIGRDGNY VMIRLPSGEM RMVHSECRAT IGVVGNGGHT DKPLGKAGRS
RWKGRWPRVR GVAMNPVDHP HGGGEGRHGG PHPVSPWGPP GRGVGTRANK RTGRFIVRRR
a) Based on an alignment of 130 L2 sequences that uniformly sample all three branches of the tree of life.
b) Sites with Shannon Entropy < 0.5 are blue (these are very highly conserved).
b) Sites of Shannon Entropy = 0 are in blue bold (these are universally conserved).
c) The ancestral peptide is underlined italic.
Figure 1. Protein L2. The amino acids are shaded by their Shannon Entropies. Blue indicates highly conserved. Red indicates variable. The ancestral peptide is indicated.
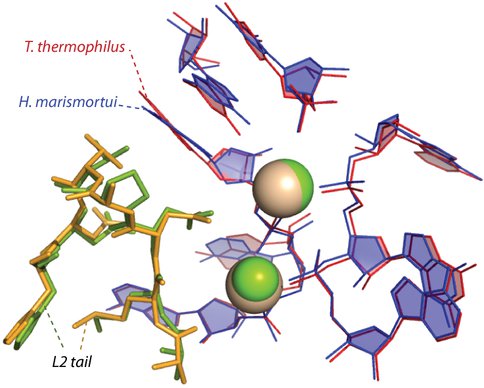
Protein L2 contains the “ancestral peptide” (Table 1, Figures 1, 2 & 3), which is defined by distinct signatures in sequence, conformation and molecular interactions. The ancestral peptide is 31 amino acids long, and contains six universally conserved amino acids, far more than any other rProtein segment of comparable length. The 12 universally conserved amino acids of L2 are primarily glycines and alanines, consistent with ancient origins 3. The ancestral peptide is the only segment of any rProtein to interact with a universally conserve magnesium ion, which together we have proposed as early ribosomal assembly components 4.
Figure 2. The central part of the ancestral peptide is highly conserved in conformation and interactions and is the only part of any ribosomal protein to interact with ancestral magnesium ions (spheres) in the core of the ribosome.
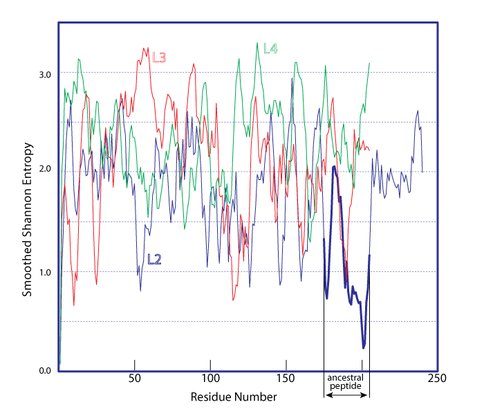
The Shannon Entropies of the ancestral peptide indicate a greater degree of conservation than for any other segment of any ribosomal protein (Figure 3, only rProteins L2, L3 and L4 are shown).
Figure 3. Sequence conservation of L2,L3,and L4. Shannon entropy for L2 (blue), L3 (red), and L4 (green) is shown as a function of residue number. The Shannon Entropies have been smoothed for clarity using a moving average filter (span=5). The residue numbers are with respect to each proteins’ alignment. Insertions are omitted. The proposed L2 ancenstral peptide is shown with a thicker blue line.
We believe this is the most conserved protein fragment in all of biology. Note that previously Gogarten and Fournier reached a different conclusion, that rProtein L4 is the oldest protein 5. Our conclusion differs from Gogarten and Fournier’s because they took statistics over entire proteins and so did not detect the special statistical signature of the ancestral peptide, which is only a portion of the non-globular tail of rProtein L2.
Recent results of Steinberg 6 have emphasized that the LSU contains information about the sequence of events in ribosomal origins and ancestral assembly. Molecular interactions within the LSU represent timing events that allow reconstruction of ancestral LSU assembly. Steinberg utilized the A minor motif to propose a model of ribosomal assembly. However, many other types of molecular interactions can be used to test and extend Steinberg’s model. For example, the A of a GNRA tetraloop frequently interacts with remote RNA elements. The implied sequence of events is tetraloop formation (first), then interaction with remote element (second). In another example, tertiary base-base interactions are comprised of a Watson-Crick pair in a stem region involved with a remote third RNA base. The implied timing sequence is base-pair formation (first), then interaction with third base (second). Our ongoing efforts are aimed at gathering and utilizing all available timing data to develop a fine-grained and cross-validated model of ribosomal evolution. We are using graph theory to automate the process and integrate a broad spectrum of input data. In addition, we have mapped the Steinberg model onto Williams’ onion model 7, showing that the onion is lower resolution, but is similar to Steinberg’s model.
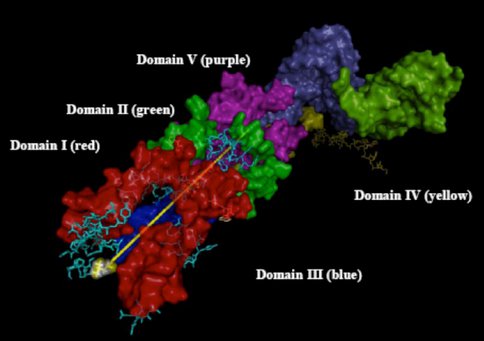
Figure 4. RNA components that form the exit tunnel (yellow line denotes path) from PTC in Domain V to the back of the 50S subunit where it exits in Domain III.
As distinct timelines for various ribosomal subsystems emerge, it is important to test for internal consistency (cross-validate). Detailed examination of the exit tunnel (Figure 4) over the past year illustrates how this can be accomplished. We assume that the tunnel began at the PTC and grew longer over time. To accommodate the growing ribosome, which is synthesizing proto-peptides of increasing length, the tunnel increased in length. Hence the time of addition of tunnel components can be correlated with the enlargement of the RNA and the various additions correlated with and in fact mapped onto a hierarchical model of 23S rRNA evolution. Thus, in the past year, the tunnel was found to pass through the Domains in the order V→IV→II→I→III, which is likely the order of addition over evolution. This order of addition is in agreement with the Steinberg model 6, the Williams onion model 7, and the Fox group’s earlier results based on connectivity 8. The mRNA channel in the 30S subunit may provide similar insights. In this case, the timing is likely bidirectional, emanating from the decoding center.
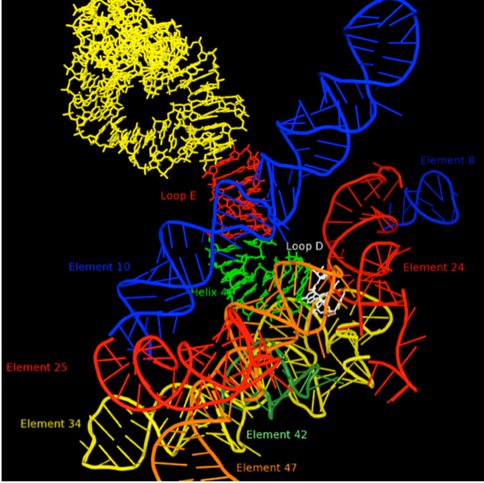
It is our view that the level of complexity that characterizes LUCA is not accidental, but rather reflects an environment when a global event in biological evolution occurred. A probable candidate for this event would be the greatly improved control of the characteristic movements of the ribosome that would be contributed by the addition of the GTPase Center to the ribosome. This addition would have accelerated the rate of protein synthesis by at least an order of magnitude and allowed major expansion of the capabilities of primitive living systems. It would have been necessary, however, to integrate the GTPase center into the inner workings of the ribosome and to do this, it is likely that a signaling system would be needed to facilitate timing of GTP cleavage. Thus, the ribosomal processes and components evolving at the LUCA boundary are likely to be of special interest. One such component is 5S rRNA, which is centrally[Insert Figure 6] located on a path, Figure 5, which ultimately connects the PTC in the 50S subunit to the decoding center in the 30S subunit. Previously a large number of 5S rRNA mutants were constructed for studies of the structure of RNA sequence space. Ongoing efforts are focused on characterizing these variants from the perspective of ribosome function. Mutations at U78 where the A-site finger crosses 5S rRNA are especially deleterious.
Figure 5. RNA environment in 50S subunit surrounding 5S rRNA Loop D (white). Other portions of the 5S rRNA are indicated in yellow, red (loop E) and green (helix 4). The various regions of 23S rRNA structure are labeled as “elements” using the nomenclature of Bokov and Steinberg, and each is colored separately. Element 10, also known as the A-site finger crosses 5S rRNA in close proximity of Loop E.
1. Fox, G. E.; Ashinikumar, K. N., The Evolutionary History of the Translation Machinery. In The Genetic Code and the Origin of Life, de Pouplana, L. R., Ed. Kluwer Academic / Plenum Publishers, New York 2004; pp 92-105.
2. Fox, G. E., Origin and evolution of the ribosome. Cold Spring Harb Perspect Biol 2010, 2 (9), a003483.
3. Lu, Y.; Freeland, S., On the evolution of the standard amino-acid alphabet. Genome Biol 2006, 7 (1), 102.
4. Hsiao, C.; Williams, L. D., A recurrent magnesium-binding motif provides a framework for the ribosomal peptidyl transferase center. Nucleic Acids Res. 2009, 37 (10), 3134-42.
5. Fournier, G. P.; Neumann, J. E.; Gogarten, J. P., Inferring the ancient history of the translation machinery and genetic code via recapitulation of ribosomal subunit assembly orders. PLoS One 2010, 5 (3), e9437.
6. Bokov, K.; Steinberg, S. V., A hierarchical model for evolution of 23S ribosomal RNA. Nature 2009, 457 (7232), 977-980.
7. Hsiao, C.; Mohan, S.; Kalahar, B. K.; Williams, L. D., Peeling the onion: ribosomes are ancient molecular fossils. Mol. Biol. Evol. 2009, 26 (11), 2415-25.
8. Hury, J.; Nagaswamy, U.; Larios-Sanz, M.; Fox, G. E., Ribosome origins: the relative age of 23S rRNA Domains. Orig Life Evol Biosph 2006, 36 (4), 421-9.
Publications
-
Bokov, K., & Steinberg, S. V. (2009). A hierarchical model for evolution of 23S ribosomal RNA. Nature, 457(7232), 977–980. doi:10.1038/nature07749
-
Fournier, G. P., Neumann, J. E., & Peter Gogarten, J. (2010). Inferring the Ancient History of the Translation Machinery and Genetic Code via Recapitulation of Ribosomal Subunit Assembly Orders. PLoS ONE, 5(3), e9437. doi:10.1371/journal.pone.0009437
-
Hsiao, C., & Williams, L. D. (2009). A recurrent magnesium-binding motif provides a framework for the ribosomal peptidyl transferase center. Nucleic Acids Research, 37(10), 3134–3142. doi:10.1093/nar/gkp119
-
Hsiao, C., Mohan, S., Kalahar, B. K., & Williams, L. D. (2009). Peeling the Onion: Ribosomes Are Ancient Molecular Fossils. Molecular Biology and Evolution, 26(11), 2415–2425. doi:10.1093/molbev/msp163
-
Smith, T. F., Lee, J. C., Gutell, R. R., & Hartman, H. (2008). The origin and evolution of the ribosome. Biol Direct, 3(1), 16. doi:10.1186/1745-6150-3-16
- Fox, G.E. & Ashinikumar, K.N. (2004). The Evolutionary History of the Translation Machinery. In: De Pouplana, L.R. (Eds.). The Genetic Code and the Origin of Life. Kluwer Academic / Plenum Publishers, New York.
- Hury, J., Nagaswamy, U., Larios-Sanz, M. & Fox, G.E. (2006). Ribosome origins: the relative age of 23S rRNA Domains. Orig Life Evol Biosph, 36(4): 421-9.
- Lu, Y. & Freeland, S. (2006). On the evolution of the standard amino-acid alphabet. Genome Biol, 7(1): 102.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Dana Cook-Schneider
Postdoc
Chiaolong Hsiao
Postdoc
Victor Stepanov
Postdoc
Jessica Bowman
Research Staff
Chad Bernier
Graduate Student
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules