2010 Annual Science Report
 Georgia Institute of Technology
Reporting | SEP 2009 – AUG 2010
Georgia Institute of Technology
Reporting | SEP 2009 – AUG 2010
High Level Theory - the Role of Mg2+ in Ribosome Assembly
Project Summary
We investigated a unique role of Mg2+ ions to form stable complexes with ribosomal RNA, and specifically their role in a formation of ancestral peptidyl transferase center using modern quantum mechanics methods. The interaction energies of ribosomal RNA with single and multiple Mg2+ cations are computed in the gas phase and water, and partitioned into specific tems. RNA-Mg interactions are compared to those with other metals, to determine why Mg2+ plays a special role in RNA folding. Additionally, we hypothesize a possible unique role of Fe2+ in a formation of ribosomal catalytic centers during early stages of life. The project is performed using NASA’s HEC supercomputer recourses.
Project Progress
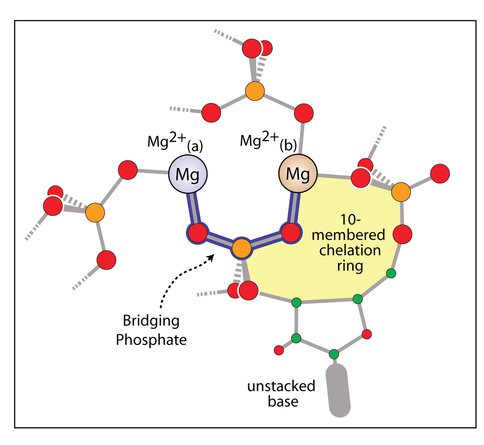
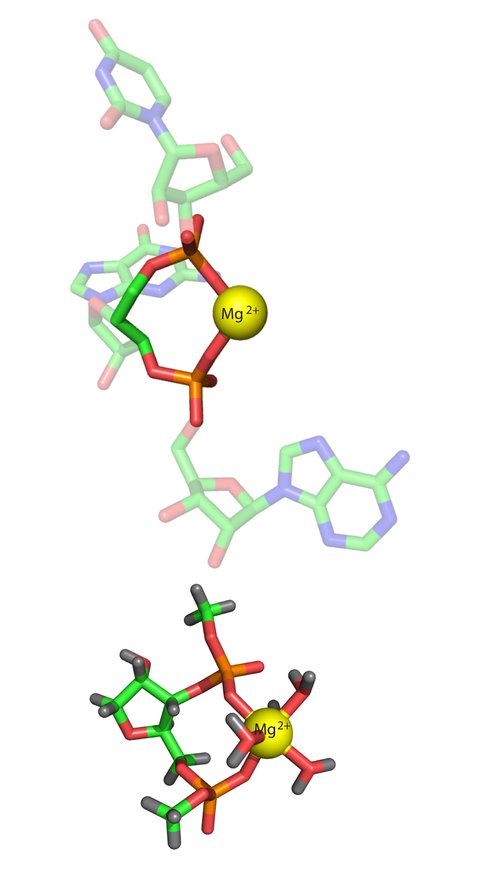
It has been shown by Hsiao and coworkers1; 2 that three Mg2+-microsclusters (Mg2+-μc’s), each containing two Mg2+ ions, provide a rigid framework surrounding the peptidyl transferase center of the ribosome (Figure 1). Bidentate RNA clamps of magnesium occur frequently in large RNAs in (Figure 2). We have found that magnesium, compared to calcium and sodium, has enhanced ability to form bidentate clamps with RNA. Bidentate RNA-sodium clamps, in particular, are unstable and spontaneously open. Due to magnesium’s size and charge density it binds more intimately than other cations to the oxyanions of RNA, so that magnesium clamps are stabilized not only by electrostatic interactions, but also by charge transfer, polarization and exchange interactions. These non-electrostatic components of the binding are quite substantial with the high charge and small interatomic distances within the magnesium complexes, but are less pronounced for calcium due to its larger size, and for sodium due to its smaller charge. Additionally, bidentate RNA clamps of magnesium are more stable than those with DNA. The source of the additional stability of RNA complexes is twofold: there is a slightly attenuated energetic penalty for ring closure in formation of RNA bidentate chelation complexes, and elevated electrostatic interactions between the RNA and cations. In sum it can be seen why sodium and calcium cannot replicate the structures or energetics of RNA-magnesium complexes. These results have been recently reported in RNA journal.
Characterization of RNA-Metal motifs through the evaluation of iron and manganese substitutes for Mg continues, as well as computations on larger cationic clusters (involving several phosphate groups and Mg2+ ions) as a way of understanding Mg2+ microclusters in a more biologically relevant context.
1. Hsiao, C., M. Tannenbaum, et al. (2008). Complexes of Nucleic Acids with Group I and II Cations. Nucleic Acid Metal Ion Interactions. N. Hud. London, The Royal Society of Chemistry: 1-35.
2. Hsiao, C. and L. D. Williams (2009). A recurrent magnesium-binding motif provides a framework for the ribosomal peptidyl transferase center. Nucleic Acids Research 37: 3134-3142.
Figure 1. A schematic diagram indicating common features of Mg2+-μc architecture. Shown is the Mg2+(a)-(O1P-P-O2P)-Mg2+(b) bridge (outlined in blue), the 10-membered ring (yellow), an unstacked base, and the additional RNA ligands that enter the Mg2+ first shell at various positions. Carbon is green, oxygen is red, and phosphorous is orange.
Figure 2. (Top) A bidentate RNA clamp of magnesium observed in the H. marismortui ribosomal LSU crystal structure (Mg 8003 from PDB entry 1JJ2). (Bottom) The RNA2—Mg2+.•(H2O)4 complex used here for QM calculations. The base has been replaced by a hydrogen atom and the chain is terminated with methyl groups.
Publications
-
Hsiao, C., & Williams, L. D. (2009). A recurrent magnesium-binding motif provides a framework for the ribosomal peptidyl transferase center. Nucleic Acids Research, 37(10), 3134–3142. doi:10.1093/nar/gkp119
- Hsiao, C., Tannenbaum, M., VanDeusen, H., Hershkovitz, E., Perng, G., Tannenbaum, A. & Williams, L.D. (2008). Complexes of Nucleic Acids with Group I and II Cations. In: Hud, N. (Eds.). Nucleic Acid Metal Ion Interactions. London: The Royal Society of Chemistry.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Anton Petrov
Postdoc
Jessica Bowman
Research Staff
Denise Enekwa
Graduate Student
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules
Objective 5.3
Biochemical adaptation to extreme environments