2010 Annual Science Report
 Georgia Institute of Technology
Reporting | SEP 2009 – AUG 2010
Georgia Institute of Technology
Reporting | SEP 2009 – AUG 2010
Fostering Synergetic Interactions Among NAI Teams Reconstructing Early Life on Earth, and Attaching a Time Scale to the Genomic Record of Life
Project Summary
Today there is a nearly universal consensus that a tree cannot describe the early evolution of life, but there is not yet a consensus about how to describe life’s early evolution. Our lab is developing new methods to incorporate symbioses and endosymbioses into reconstructions of early life on Earth and thereby represent life as a combination of tree like- and symbiotic like- evolution. Using these improved methods, we are attaching a time scale to the rings that represent this early evolution, in order to better understand significant early events in Earth’s history like the origin of oxygenic photosynthesis.
Project Progress
Today it is widely agreed that a tree cannot describe the early evolution of life1, but there is not yet a consensus about how to describe life’s early evolution. Recent work suggests a new way for describing the progression of early life by incorporating symbioses and endosymbioses into our reconstructions of life 2. This work provides evidence that the Gram negative bacteria with double cell membranes originated as the result of an endosymbiosis between prokaryotes (as opposed to Gram positive bacteria, which have a single cell membrane). Hence the evolutionary path of the double membrane group is represented by a specific ring, rather than by a tree (Figure 1). Gram negative bacteria, which include the Cyanobacteria and other intriguing prokaryotes, have profoundly altered life on Earth by producing 20% of the Earth’s atmosphere. Knowledge of the specific ring topology that accounts for the emergence of Gram negative bacteria allows us to determine when particular branches within the ring formed. This makes it possible to attach a reliable time scale to branching within the “tree of life”, and thereby reduce the errors that were generated by using trees, rather than rings, for dating. For example, photosynthesis originated at the base of the yellow branch that connects the Clostridia and the Double Membrane prokaryotes in the Ring above. Trees have previously been unable to pinpoint this site because the tree of prokaryotes is highly unresolved 3. Rings can now resolve this location and others, allowing us to begin to attach reliably a molecular clock.
In 2010, a Strategic Planning-based winter meeting was held at UCLA for NAI team members from diverse Earth-related sciences broadly related to this proposal. This meeting was extremely successful, and the cross-disciplinary interactions continued both in and out of the session (see the pictures below). As a result, productive discussions regarding how to date the evolution of life on Earth were initiated, and potential collaborations were explored. These will become important contacts for future studies.
We are currently focusing on understanding the phylogenetic events (speciation, symbioses, and possible endosymbioses) that accompanied the beginnings of oxygenic photosynthesis. The rise of oxygen in Earth’s atmosphere is one of the better dated geological landmarks, and the importance of oxygen for the emergence of eukaryotic life makes it a pivotal time calibration point, that must be related to the tree/rings of life.
A UCLA undergraduate, Arnold Pham, has been making great progress working on finding better ways to access sequence information that is central for reconstructing the historical past. He is developing software for downloading phylogenetically useful data, and is starting to automate methods for reconstructing rings 4 from gene presence/absence data.
1. Doolittle, W. F., Phylogenetic classification and the universal tree. Science 1999, 284 (5423), 2124-2128.
2. Lake, J. A., Evidence for an early prokaryotic endosymbiosis. Nature 2009, 460 (7258), 967-971.
3. Ludwig, W.; Klenk, H. P., Overview: a phylogenetic backbone and taxonomic framework for procaryotic systematics. In Bergey’s Manual of Systematic Bacteriology, Boone, D. R.; Castenholz., R. W., Eds. Springer: New York, Berlin, 2001; Vol. 1, pp 49-65.
4. Lake, J. A., Reconstructing evolutionary graphs: 3D parsimony. Molecular Biology and Evolution 2008, 25 (8), 1677-1682.
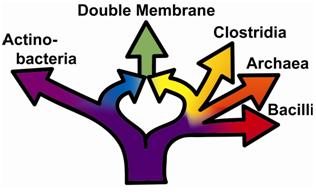
Figure 1. Evolutionary path represented by a specific ring, rather than by a tree
Publications
-
Lake, J. A. (2008). Reconstructing Evolutionary Graphs: 3D Parsimony. Molecular Biology and Evolution, 25(8), 1677–1682. doi:10.1093/molbev/msn117
-
Lake, J. A. (2009). Evidence for an early prokaryotic endosymbiosis. Nature, 460(7258), 967–971. doi:10.1038/nature08183
- Doolittle, W.F. (1999). Phylogenetic classification and the universal tree. Science, 284(5423): 2124-2128. doi:10.1126/science.284.5423.2124
- Ludwig, W. & Klenk, H.P. (2001). Overview: a phylogenetic backbone and taxonomic framework for procaryotic systematics. In: Boone, D.R. & Castenholz., R.W. (Eds.). Bergey’s Manual of Systematic Bacteriology. Vol. 1. New York, Berlin: Springer.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Arnold Pham
Undergraduate Student
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules
