2007 Annual Science Report
 Marine Biological Laboratory
Reporting | JUL 2006 – JUN 2007
Marine Biological Laboratory
Reporting | JUL 2006 – JUN 2007
Microbial Diversity and Population Structure Studies in the Rio Tinto
Project Summary
Project Progress
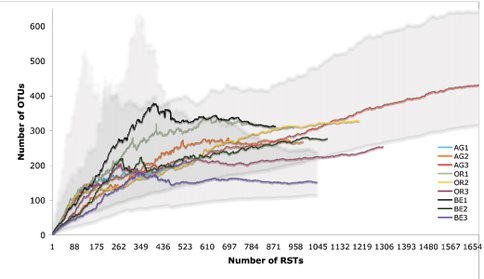
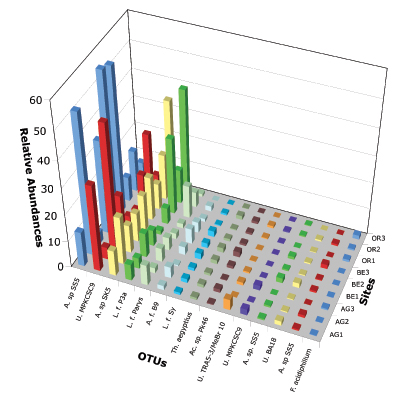
A major synthesis of our SARST-V6 data are included in a manuscript entitled: “The Rio Tinto as a model system for studying microbial evolutionary ecology” to be submitted for publication later this summer. In this study we use a relatively new tag sequencing method, Serial Analysis of Ribosomal Sequence Tags of the V6 hypervariable region (SARST-V6) coupled with fine measurements of physico-chemical parameters to explore how ecological and evolutionary factors influence patterns of bacterial community structure emerging at the most extreme stations of the Rio Tinto. Through the integration of a careful experimental design that includes sample replication and high-throughput molecular methods, we deeply explore the bacterial diversity in the Rio Tinto and its link to similar acidic environments. We show the importance of accurate alpha and beta ecological diversity measurements, which, coupled with advanced geochemical and ecological analysis tools, reveal evolutionarily ecological units of neutrally divergent microdiverse ribotypes that behave as ecotypes intimately linked to their environment. Figure 1 shows a histogram plot of the major bacterial operational taxonomic units present in the river. Figure 2 depicts the variation of non-parametric richness estimator Chao 1 with sampling effort for the RT sites. The curve levels off in all but two of our sites AG1 and AG3 indicating that our sampling efforts have reached saturation in most cases.
Collection and curation of our three-domain sequence data targeting the V4 through V8 regions of the small-subunit rRNA gene are complete. Analyses have begun to reveal very interesting lineage-specific patterns of diversity. We have recovered over 7,000 sequences as part of this study. As with the SARST-V6 data, these data will provide both species diversity and relative abundance information, as well as reveal relative abundance data for each domain within our samples. We intend to submit the results of this work in a manuscript by the end of this year.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Linda Amaral Zettler
Co-Investigator
Maria Aguilera
Collaborator
Ricardo Amils
Collaborator
Carmen Palacios
Collaborator
Erik Zettler
Collaborator
Abby Laatsch
Research Staff
Susanna Theroux
Research Staff
-
RELATED OBJECTIVES:
Objective 5.1
Environment-dependent, molecular evolution in microorganisms
Objective 5.2
Co-evolution of microbial communities