2006 Annual Science Report
 Marine Biological Laboratory
Reporting | JUL 2005 – JUN 2006
Marine Biological Laboratory
Reporting | JUL 2005 – JUN 2006
Genome-Genome Integration: Symbiosis, Genetic Assimilation, and Evolutionary Innovation
Project Summary
The broad goal of this project is to clarify genetic changes that mediate the establishment and diversification of genome-genome interactions. Using insect-associated endosymbionts as model systems, we are examining the molecular and evolutionary forces that shape associations between bacteria and eukaryotic hosts.
Project Progress
The broad goal of this project is to clarify genetic changes that mediate the establishment and diversification of genome-genome interactions. Using insect-associated endosymbionts as model systems, we are examining the molecular and evolutionary forces that shape associations between bacteria and eukaryotic hosts. Our studies examine Proteobacterial species that represent both long-term, stable mutualisms and more dynamic, parasitic interactions.
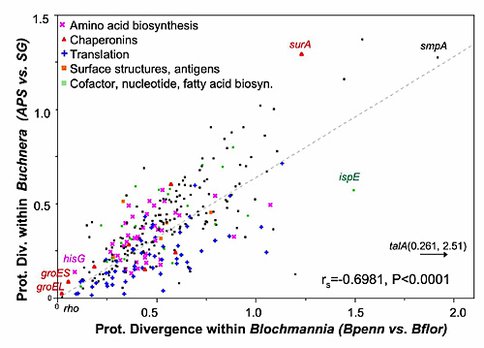
Our completion of the 792-kb genome of Blochmannia, an obligate endosymbiont of various ant species (Degnan et al. 2005), provided the basis for new comparative studies of forces shaping sequence evolution and genome architecture in intracellular bacteria. For example, our analyses of protein evolution revealed 100-fold variation in amino acid substitution rates among loci, indicating variable functional constraints across mutualist functions (Fry and Wernegreen 2005; Degnan et al. 2005; manuscript in preparation). We also found accelerated rates of protein evolution in endosymbionts and correlated divergence levels among orthologs shared by mutualist groups, highlighting the predictable consequences of long-term intracellularity on patterns of protein evolution.
Our current work also employs functional genomic approaches to explore how bacteria respond to the dynamic environment of animal hosts. Preliminary data indicate functional plasticity in the ant-bacterial interaction, as key symbiotic loci vary in expression across distinct host developmental stages. Functional studies also revealed that transcriptional slippage influences expression of symbiont genes with long homopolymeric tracts (9-12 consecutive A’s or T’s). Finally, our comparisons of gene contents among mutualist groups are distinguishing convergent patterns of gene loss in response to an intracellular lifestyle versus genomic differences that catalyze specific host interactions (Wernegreen 2005; Wernegreen submitted).
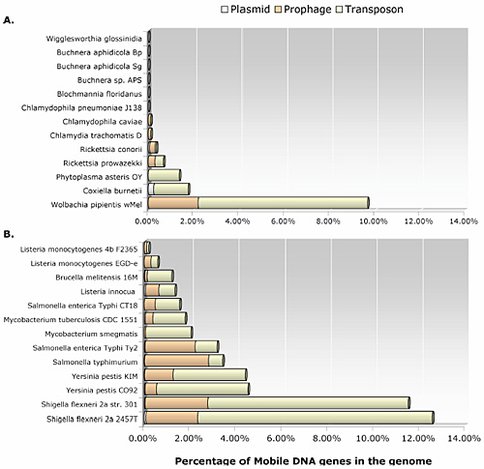
This year we continued our genomic and molecular studies on Wolbachia endosymbionts – one of the most widespread groups of infectious bacteria in the terrestrial biosphere. These inherited microbes infect the germline tissues of arthropods and nematodes, are closely related to the bacterial ancestor of mitochondria, and show both parasitic and mutualistic associations within a wide invertebrate host range. We use these features to study the evolutionary forces guiding some microbial symbionts into organelles and others into long-term parasites. This past year, we published seven articles and have another four manuscripts currently in review. Among our most exciting findings, we reported in PLoS Pathogens this past May that arthropods, Wolbachia, and a horizontally-transferring bacteriophage of Wolbachia form a set of tritrophic interactions in which all three members are integral to understanding this important, heritable symbiosis (Bordenstein et al. 2006). This work was featured by several scientific journals including The Scientist. Our comprehensive review article on mobile DNA in obligate intracellular bacteria (featured in a Nature Reviews special issue on horizontal gene transfer) highlights the extraordinary phage and transposon colonization of the Wolbachia genome relative to other obligate and facultative intracellular species (Bordenstein and Reznikoff 2005).
Bioinformatic and molecular phylogenetic studies this past year determined that Wolbachia have highly fluid and diverse genomes due to intragenic recombination (Baldo et al. 2006), a pattern atypical of other obligate intracellular symbionts. Owing to the genetic recombination, micro- and macro-level taxonomy is therefore locus independent. A large-scale sequencing regime allowed us to create a multi-locus sequence typing system for this bacterium (Baldo et al. 2006; Paraskevopoulos et al. 2006; Casiraghi et al. 2005). We are learning through our research progress from this award that the success of this endosymbiosis across the planet is associated with extraordinary genome fluidity via recombination and mobile DNA insertions/deletions.
Mutualists:
- Comparisons among endosymbiont genomes revealed similar shifts in rates and patterns of protein evolution. Intracellular bacteria show significant elevation in evolution rates across nearly all loci and exhibit parallel divergences among shared genes (Fry and Wernegreen 2005; Degnan et al. 2005; manuscript in preparation). These patterns indicate predictable consequences of long-term intracellularity on protein divergences.
- Bacteria engaged in long-term associations with eukaryotic host cells show convergent patterns of gene loss in response to an intracellular lifestyle. Exceptions to this trend highlight genomic differences that catalyze specific host interactions, such as the retention of specific biosynthetic loci that match host dietary requirements (Wernegreen 2005; Wernegreen submitted).
Wolbachia:
- Quantitative and electron microscopic analyses showed that the binary interactions of obligate Wolbachia symbionts and eukaryotes can be perturbed by Life’s most creative evolutionary force on the planet — bacteriophages
(Fig. 3).
Our studies specify that bacteriophages, for instance, regulate within-host densities and phenotypes of the Wolbachia community within invertebrates (Bordenstein et al, 2006). These findings suggest that bacteriophage are not only important to microbial community composition in large open environments, but also to the closed intracellular niches of some long-term bacterial symbionts of eukaryotes. Complex life can thus be shaped by the tripartite genomes of phage, bacteria, and eukaryotes. - Comparative genome analyses among the olbigate intracellular bacteria showed that the majority of species harbor active and inactive remnants of mobile DNA, including plasmids, transposons, and prophage (Bordenstein and Reznikoff, 2005). Such genomes are commonly thought to be streamlined of parasitic genetic elements and impervious to lateral gene acquisitions. Our findings suggest that the cells of complex eukaryotes can serve an incubator for gene transfer and recombination among bacterial species that coinfect eukaryotic hosts.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Seth Bordenstein
Co-Investigator
John Werren
Collaborator
Sarah Biber
Research Staff
Seth Kauppinen
Research Staff
Adam Lazarus
Research Staff
Michelle Marshall
Research Staff
-
RELATED OBJECTIVES:
Objective 4.2
Foundations of complex life
Objective 5.1
Environment-dependent, molecular evolution in microorganisms
Objective 5.2
Co-evolution of microbial communities