Georgia Institute of Technology
10/2016 -
12/2019
(CAN 7)
Reliving the Past
(Note: The team was previously based at the University of Montana, Missoula.)
Not so long ago a young man wandered along the Down Bank in Kent and wondered at the beauty of a hillock called Orchis Bank that was teeming with life. Out of his wandering, and all the years of wandering and wondering that preceded it, a grand idea took shape, the idea that all creatures in view and within earshot, “so different from each other, and dependent upon each other, had all been produced by laws acting around us.” To discover the laws that create Darwin’s “tangled bank” remains one of biology’s grand challenges, one that requires understanding how differences among forms are selected for and how interdependence among forms is enforced. In nature, meeting this challenge is complicated by the fact that selection often varies widely over space and through time. Further, in nature we are slaves of time, forced to consider the consequences of evolution as we seek to infer its causes. Laboratory evolution experiments, especially those using microbes, offer an attractive way to meet this challenge. Because microorganisms can be cryopreserved, we can accumulate a frozen fossil record of replicated experiments as we study, under controlled conditions, the dynamic interplay between genotype and phenotype, and the interactions among phenotypes in simple communities. Laboratory evolution experiments make it possible to discover why evolution takes certain paths and not others in a universe of limitless possibilities. Using experimental microbial evolution we can revisit life’s possibilities. Moreover, because the principal actors in the first 2.5 billion years of life’s drama were microbes, we are obliged to use these forms to unlock the secrets of how life underwent transitions from simple autocatalytic units to self-organizing, self-replicating systems that range in complexity from single cells to ecosystems.
The Georgia Institute of Technology team (previously based at the University of Montana, Missoula) has assembled an interdisciplinary group of investigators to address, using experimental microbial genomics, this overarching question: What forces bring about major transitions in the evolution of biocomplexity? This team’s program of study aims to meet the grand challenge of illuminating and interpreting the laws that produce the “tangled bank” so that we may better understand and appreciate the “grandeur in this view of life.”
Darwin’s grand view was one in which the “Struggle for Life” figured prominently. And indeed, the metaphor of “nature red in tooth and claw” still looms large in the imagination of laymen and scientists alike. However, for decades now it has been recognized that not just competitive, but also cooperative interactions are fundamental features of biological systems ranging from enzymes to organelles, cells and societies of cells and organisms. The Georgia Tech team is organized around five questions related to major transitions in the history of Life: (1) How do enzymes and metabolic networks evolve? (2) How did the eukaryotic cell come to be, specifically the cell that contained a mitochondrion? (3) How do symbioses arise? (4) How does multicellularity evolve? and (5) How do pleiotropy, epistasis and mutation rate constrain the evolution of novel traits? A unifying theme underlying these questions is: how do cooperative vs. competitive interactions play out in driving major transitions that occur when independently replicating entities combine into a larger, more complex whole? This NAI team seeks to understand the general principles that are likely to hold wherever life exists and to collectively affect its degree of complexity.
The Georgia Tech team addresses its five overarching questions via eight integrated research activities that share well established biochemical, genetic and genomic approaches, some of which were pioneered by members of this consortium. Concerning Question 1, on the evolution of metabolism, this team will explore how fitness can be improved in a microbe whose growth rate is limited by an inefficient metabolic enzyme (Project 1 From generalist to specialists (or not)), and will seek to discover how extant symbiont enzymes may have contributed to genome reduction by reverting to the characteristics of ancestral generalist enzymes. (Project 2 Function by reduction). These activities complement those of Project 3 (Consequences of recA duplication) which will elucidate the impact of duplication of the bacterial recombinase gene on genome evolution and fitness.
Concerning Question 2, the mechanisms of eukaryogenesis, this team will investigate how the mitochondrial endosymbiosis might have arisen by selection for cooperation between two bacteria that initially had a predator-prey relationship (Project 4 A model for eukaryogenesis: Coevolution of E. coli and its parasite B. bacteriovorus). This research dovetails with Project 2 on the evolution of endosymbionts, a trait observed in nearly every eukaryotic phylum.
Regarding Question 3, symbiosis, the objectives and results of Project 4 integrate with that of Project 5: Adaptation, mutation supply, and evolution of synergy, which seeks to unravel the genetic and ecological factors driving cooperative interactions among cells in biofilms. Such interactions bear critically on Question 4, multicellularity, the genetics and evolution of which is to be studied by experimentally evolving this trait in a single-celled green alga (Project 6: The evolution of complexity via multicellularity and cell differentiation).
Finally, regarding Question 5, constraints on the evolution of novelty, this team will investigate how the interdependency of mutation and recombination ensures the persistence of populations and conditions that drive transitions to cooperation and multicellularity (Project 7: Error rate and the origin and early evolution of Life), and will use innovative molecular barcoding techniques first to identify beneficial mutations that occur as yeast adapt to resource limitation and to determine which interactions forbid certain trajectories and which open up others. (Project 8: Limits to optimality in adaptive evolution). These empirical studies will be further integrated by theoretical investigations on epistasis, cooperation and biocomplexity.
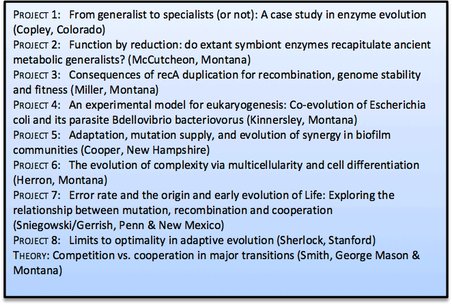 Summary of research projects of the Georgia Tech team, with lead investigators.
Summary of research projects of the Georgia Tech team, with lead investigators.
Annual Reports
TEAM LOGO:

-

Frank Rosenzweig
-
Active Dates:
10/2016 - 12/2019 -
Team Website:
Coming Soon -
Members:
55 (See All)


