The majority of life’s history has been dominated by microbes whose metabolic inventions have significantly altered the Earth’s surface environment and, in turn, impacted the evolution of life on Earth. Because unicellular microorganisms do not readily leave diagnostic morphological fossils, alternative strategies are necessary for studying microbial communities in the context of the Earth’s distant past. One predominant strategy is to correlate organic compounds deposited by ancient microbes and preserved in sedimentary rock with lipid molecules produced by modern microorganisms. One such class of molecular fossils are the hopanes, pentacyclic triterpenoid lipids that are clearly the diagenetic products of bacterial hopanoid lipids. However, very little is known about the biosynthesis and physiological function of hopanoids in extant bacteria and this in turn has made it difficult to properly interpret the occurrence of hopane signatures in the rock record.
To address this problem, we have begun to decipher the biosynthesis and function of hopanoids in methanotrophic bacteria. Using comparative genomics and gene deletion analysis, we have identified genes involved in hopanoid biosynthesis. Bioinformatics analyses of these biosynthesis genes have demonstrated that the taxonomic distribution of hopanoid producing bacteria is much more diverse than previously thought. Phenotypic analysis of hopanoid biosynthesis mutants revealed a role for these molecules in maintaining outer membrane impermeability as well as a potential role in late stationary phase survival. Taken together, these results suggest that the occurrence of hopanoids in ancient environments may better reflect a certain bacterial response (i.e. bacterial stress) rather than a specific bacterial taxa or metabolism.
 Investigating Habitable Environments on Mars Using Orbital and Rover-Based Imaging Spectroscopy
Investigating Habitable Environments on Mars Using Orbital and Rover-Based Imaging Spectroscopy Chemical Gardens, Chimneys, and Fuel Cells: Simulating Prebiotic Chemistry in Hydrothermal Vents on Ocean Worlds
Chemical Gardens, Chimneys, and Fuel Cells: Simulating Prebiotic Chemistry in Hydrothermal Vents on Ocean Worlds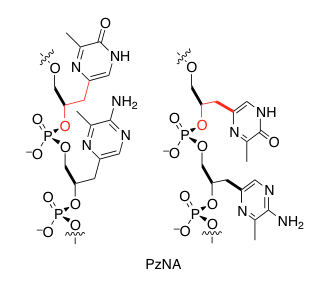 The Synthesis of an Artificial Genetic Polymer: From Small Molecules to Proto-Nucleic Acids
The Synthesis of an Artificial Genetic Polymer: From Small Molecules to Proto-Nucleic Acids Quantifying Constraints on Metabolic Diversity Patterns
Quantifying Constraints on Metabolic Diversity Patterns