
Deeply buried marine sediments may house a large amount of the Earth’s microbial population. Initial studies based on 16S rRNA clone libraries suggest that these sediments contain unique phylotypes of microorganisms, particularly from the archaeal domain. Since this environment is so difficult to study, microbiologists are challenged to find ways to examine these populations remotely. A major approach taken to study this environment uses massively parallel sequencing to examine the inner genetic workings of these microorganisms after the sediment has been drilled. Both metagenomics and tagged amplicon sequencing have been employed on deep sediments, and initial results show that different geographic regions can be differentiated through genomics and also minor populations may cause major geochemical changes.
 Investigating Habitable Environments on Mars Using Orbital and Rover-Based Imaging Spectroscopy
Investigating Habitable Environments on Mars Using Orbital and Rover-Based Imaging Spectroscopy Chemical Gardens, Chimneys, and Fuel Cells: Simulating Prebiotic Chemistry in Hydrothermal Vents on Ocean Worlds
Chemical Gardens, Chimneys, and Fuel Cells: Simulating Prebiotic Chemistry in Hydrothermal Vents on Ocean Worlds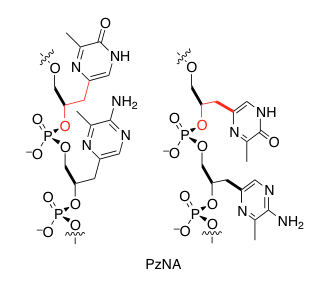 The Synthesis of an Artificial Genetic Polymer: From Small Molecules to Proto-Nucleic Acids
The Synthesis of an Artificial Genetic Polymer: From Small Molecules to Proto-Nucleic Acids Quantifying Constraints on Metabolic Diversity Patterns
Quantifying Constraints on Metabolic Diversity Patterns