2015 Annual Science Report
 University of Wisconsin
Reporting | JAN 2015 – DEC 2015
University of Wisconsin
Reporting | JAN 2015 – DEC 2015
Project 1D: Comparative Genomic Analysis of Chemolithotrophic Fe(II)-Oxidizing Bacteria
Project Summary
A comparative genomic analysis was performed to identify candidate genes involved in extracellular electron transfer (EET) by Fe(II)-oxidizing bacteria (FeOB). The analysis included a variety of publically-available FeOB genomes, together with genomes from FeOB isolated from subsurface sediments, previously-isolated marine basalt-associated FeOB, and metagenomes from chemolithoautotrophic aerobic pyrite-oxidizing and nitrate-reducing Fe(II)-oxidizing enrichment cultures. We identified outer membrane multi-copper oxidase (MCO) genes homologous to proteins known to be involved in EET in several of the FeOB genomes, as well as homologs to the outer membrane c-type cytochrome (ctyc) Cyc2 known to be involved in bacterial Fe(II) oxidation by Acidithiobacillus ferrooxidans under acidic conditions. Further, we found gene clusters that may potentially encode novel “porin-cytochrome-c protein complex” (PCC) in the well-known neutral-pH FeOB S. lithotrophicus ES-1, and homologous operons were found in other recognized FeOB (Leptothrix cholodnii SP-6 and Leptothrix ochracea L12. Another gene cluster consisting of a porin and three periplasmic multiheme cytc was identified in Hyphomicrobium sp. genome retrieved from a pyrite-oxidizing enrichment culture, and its homologous gene clusters are also present in five marine Zetaproteobacterial FeOB genomes. Overall, this analysis, which is based on our current understanding of bacterial EET in Fe redox reactions, provides a list of candidate genes for further experimental and genomic studies.
Project Progress
The goal of this project is to better understand electron transfer and genes involved in bacterial Fe(II) oxidation at circumneutral pH through comparative genomic analysis. Identification of the functional biological components (i.e. genes and proteins) involved in these chemolithotrophic processes is critical for making a concrete linkage between biological metabolism and the generation of mineralogical and isotopic biosignatures in relation to redox gradients on Earth and other rocky planets.
We have obtained genomes from Fe(II)-oxidizing bacteria (FeOB) isolated from subsurface sediments (Benzine et al., 2013) and previously-isolated marine basalt-associated FeOB, as well as metagenomes of a chemolithoautotrophic pyrite-oxidizing enrichment culture (Percak-Dennett et al., 2016) and a chemolithoautotrophic nitrate-dependent Fe(II)-oxidizing enrichment cultures (Straub et al., 1996; He et al., 2016). Together with the increasing number of publically available FeOB genomes, we performed a comparative analysis to search for candidate Fe(II) oxidase genes involved in extracellular electron transfer (EET) by FeOB.
The redox active enzymes in iron oxidation and reduction are often c-type cytochromes (cytc) (Richter et al., 2012; Shi et al., 2012) and to a lesser extent, multicopper oxidases (MCO) (Mehta et al., 2006). Based on current knowledge of EET, we generalized the Gram-negative bacterial iron oxidase (and reductase) systems into two models:
In one model, the redox active enzyme is secreted as an outer membrane or extracellular protein in order to directly interact with extracellular iron. Examples include the outer membrane c-type cytochrome (cytc) Cyc2 in acidiphilic Fe(II)-oxidizer Acidithiobacillus ferroxidans (Castelle et al., 2008), and outer membrane multiheme cytc (e.g. OmcB, OmcC, OmcE and OmcS) (Leang et al., 2003; Mehta et al., 2005) and MCO (e.g. OmpB and OmpC) (Mehta et al., 2006; Holmes et al., 2008) in Fe(III)-reducing Geobacter sulfurreducens.
In the other model, the redox active enzyme, which is often a multiheme cytc, is secreted to the periplasm, and embedded into a porin on the outer membrane to directly or indirectly interact with extracellular iron, and this system was referred to as the “porin-cytochrome c protein complex” (PCC) (Richardson et al., 2012). Examples include the homologous MtrAB in Shewanella oneidensis (Beliaev and Saffarini, 1998), PioAB in Rhodopseudomonas palustris TIE-1 (Jiao and Newman, 2007) and MtoAB in Sideroxydans lithotrophicus ES-1 (Liu et al., 2012; Shi et al., 2012; Emerson et al., 2013), as well as the more recently discovered novel PCC in G. sulfurreducens (Liu et al., 2014).
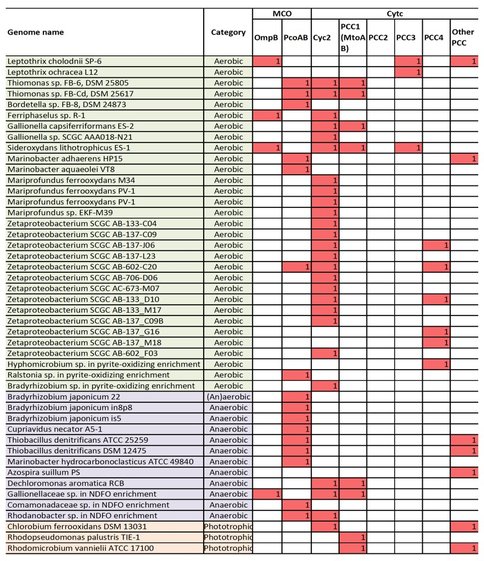
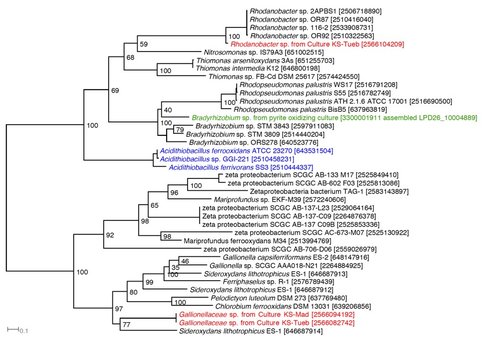
We therefore searched for genes that encode outer-membrane/extracellular cytc or MCO, and gene clusters that potentially encode porin-periplasmic cytc complex (i.e. PCC) or porin-periplasmic MCO complex in neutrophilic FeOB, including microaerophilic, anaerobic (nitrate-reducing), and phototrophic FeOB. We identified outer membrane MCO homologous to OmpB in several FeOB, and outer membrane Cyc2 in many microaerophilic FeOB and several anaerobic or phototrophic FeOB (Figure 1). Notably, these Cyc2 proteins are distant homologs of Cyc2 in A. ferroxidans (Figure 2). PCC genes homologous to MtrAB/MtoAB/PioAB (tentatively referred to as “PCC1”) were identified in a few microaerophilic, anaerobic, or phototrophic FeOB, while PCC homologous to G. sulfurreducens (Liu et al., 2014) (tentatively “PCC2”) is not present in any of the FeOB.
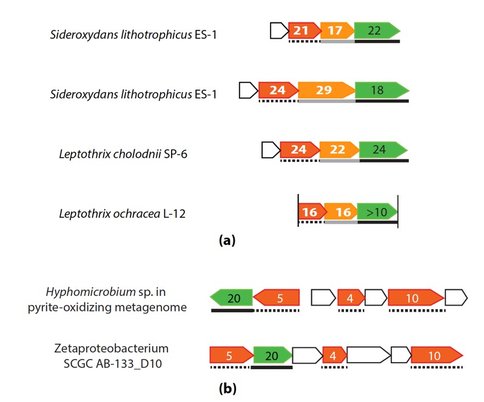
Further, we found gene clusters that may potentially encode novel PCCs. For example, two operons, each consisting of a porin, an extracellular multiheme cytc and a periplasmic multiheme cytc (tentatively “PCC3”) were identified in S. lithotrophicus ES-1, and homologous operons were found in Leptothrix cholodnii SP-6 and Leptothrix ochracea L12 (Figure 3a). Another gene cluster consisting of a porin and three periplasmic multiheme cytc (tentatively “PCC4”) was identified in hyphomicrobium sp. genome retrieved from a pyrite-oxidizing enrichment culture (Figure 3b), and its homologous gene clusters are also present in five marine Zetaproteobacterial FeOB genomes (Figure 1).
In addition to putative PCC, operons consisting of porin and periplasmic MCO that are homologous to PcoAB genes were also identified in a number of FeOB (Figure 1). Notably, we did not find any obvious candidate Fe(II) oxidase genes in about half of anaerobic FeOB genomes searched, probably suggesting that their electron transfer mechanisms in Fe(II) oxidation might be different from the two general models described above. Overall, this analysis, which is based on our current understanding of bacterial EET in iron redox reactions, provides a list of candidate genes for further experimental and genomic studies.
References
Alneberg, J., B. S. Bjarnason, I. de Bruijn, M. Schirmer, J. Quick, U. Z. Ijaz et al. 2014. Binning metagenomic contigs by coverage and composition. Nat. Methods 11:1144-1146.
Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402.
Caporaso, J. G., J. Kuczynski, J. Stombaugh, K. Bittinger, F. D. Bushman, E. K. Costello et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335-336.
Grotzinger, J. P., D. Y. Sumner, L. C. Kah, K. Stack, S. Gupta, L. Edgar et al. 2014. A habitable fluvio-lacustrine environment at Yellowknife Bay, Gale Crater, Mars. Science 343:DOI: 10.1126/science.1242777.
He, S., C. Tominski, A. Kappler, S. Behrens, and E. E. Roden. 2016. Metagenomic analyses of the autotrophic Fe(II)-oxidizing, nitrate-reducing enrichment Culture KS. Appl Environ Microb In press.
Kozubal, M. A., W. P. Inskeep, and R. E. Macur. 2012. Geomicrobiology of iron oxide mats from acidic geothermal springs in Yellowstone National Park: Microbial community structure, isolation of novel species and mechanisms of iron oxidation. Front. Microbiol. 3:109.
Markowitz, V. M., E. Szeto, K. Palaniappan, Y. Grechkin, K. Chu, I. M. A. Chen et al. 2007. The integrated microbial genomes (IMG) system in 2007: data content and analysis tool extensions. Nucl. Acids Res.gkm846.
Percak-Dennett, E. M., and E. E. Roden. 2014. Geochemical and microbiological responses to oxidant introduction in reduced subsurface sediment from the Hanford 300 Area, WA. Environ. Sci. Technol. 48:9197-9204.
Percak-Dennett, E. M., S. He, B. J. Converse, H. Konishi, H. Xu, C. S. Chan et al. 2016. Aerobic microbial pyrite oxidation at circumneutral pH. Manuscript in preparation.
Podosokorskaya, O. A., V. V. Kadnikov, S. N. Gavrilov, A. V. Mardanov, A. Y. Merkel, O. V. Karnachuk et al. 2013. Characterization of Melioribacter roseus gen. nov., sp nov., a novel facultatively anaerobic thermophilic cellulolytic bacterium from the class Ignavibacteria, and a proposal of a novel bacterial phylum Ignavibacteriae. Environ. Microbiol. 15:1759-1771.
Wang, Q., G. M. Garrity, J. M. Tiedje, and J. R. Cole. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261-5267.
Weiss, J. V., J. A. Rentz, T. Plaia, S. C. Neubauer, M. Merrill-Floyd, T. Lilburn et al. 2007. Characterization of neutrophilic Fe(II)-oxidizing bacteria isolated from the rhizosphere of wetland plants and description of Ferritrophicum radicicola gen. nov. sp. nov., and Sideroxydans paludicola sp. nov. Geomicrobiol. J. 24:559-570.
Wrighton, K. C., B. C. Thomas, I. Sharon, C. S. Miller, C. J. Castelle, N. C. VerBerkmoes et al. 2012. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337:1661-1665.
Wrighton, K. C., C. J. Castelle, M. J. Wilkins, L. A. Hug, I. Sharon, Beliaev, A.S., and Saffarini, D.A. (1998) Shewanella putrefaciens mtrB encodes an outer membrane protein required for Fe(III) and Mn(IV) reduction. J Bacteriol 180: 6292-6297.
Benzine, J., Shelobolina, E., Xiong, M.Y., Kennedy, D.W., McKinley, J.P., Lin, X., and Roden, E.E. (2013) Fe-phyllosilicate redox cycling organisms from a redox transition zone in Hanford 300 Area sediments. Frontiers in Microbiology 4: 388.
Castelle, C., Guiral, M., Malarte, G., Ledgham, F., Leroy, G., Brugna, M., and Giudici-Orticoni, M.T. (2008) A new iron-oxidizing/O2-reducing supercomplex spanning both inner and outer membranes, isolated from the extreme acidophile Acidithiobacillus ferrooxidans. J Biol Chem 283: 25803-25811.
Emerson, D., Field, E.K., Chertkov, O., Davenport, K.W., Goodwin, L., Munk, C. et al. (2013) Comparative genomics of freshwater Fe-oxidizing bacteria: implications for physiology, ecology, and systematics. Front Microbiol 4: 254.
Holmes, D.E., Mester, T., O’Neil, R.A., Perpetua, L.A., Larrahondo, M.J., Glaven, R. et al. (2008) Genes for two multicopper proteins required for Fe(III) oxide reduction in Geobacter sulfurreducens have different expression patterns both in the subsurface and on energy-harvesting electrodes. Microbiology 154: 1422-1435.
Jiao, Y., and Newman, D.K. (2007) The pio Operon Is Essential for Phototrophic Fe(II) Oxidation in Rhodopseudomonas palustris TIE-1. Journal of Bacteriology 189: 1765-1773.
Leang, C., Coppi, M.V., and Lovley, D.R. (2003) OmcB, a c-type polyheme cytochrome, involved in Fe(III) reduction in Geobacter sulfurreducens. J Bacteriol 185: 2096-2103.
Liu, J., Wang, Z., Belchik, S.M., Edwards, M.J., Liu, C., Kennedy, D.W. et al. (2012) Identification and Characterization of MtoA: A Decaheme c-Type Cytochrome of the Neutrophilic Fe(II)-Oxidizing Bacterium Sideroxydans lithotrophicus ES-1. Front Microbiol 3: 37.
Liu, Y., Wang, Z., Liu, J., Levar, C., Edwards, M.J., Babauta, J.T. et al. (2014) A trans-outer membrane porin-cytochrome protein complex for extracellular electron transfer by Geobacter sulfurreducens PCA. Environ Microbiol Rep 6: 776-785.
Mehta, T., Coppi, M.V., Childers, S.E., and Lovley, D.R. (2005) Outer membrane c-type cytochromes required for Fe(III) and Mn(IV) oxide reduction in Geobacter sulfurreducens. Appl Environ Microbiol 71: 8634-8641.
Mehta, T., Childers, S.E., Glaven, R., Lovley, D.R., and Mester, T. (2006) A putative multicopper protein secreted by an atypical type II secretion system involved in the reduction of insoluble electron acceptors in Geobacter sulfurreducens. Microbiology 152: 2257-2264.
Richardson, D.J., Butt, J.N., Fredrickson, J.K., Zachara, J.M., Shi, L., Edwards, M.J. et al. (2012) The ‘porin-cytochrome’ model for microbe-to-mineral electron transfer. Mol Microbiol 85: 201-212.
Richter, K., Schicklberger, M., and Gescher, J. (2012) Dissimilatory Reduction of Extracellular Electron Acceptors in Anaerobic Respiration. Applied and Environmental Microbiology 78: 913-921.
Shi, L., Rosso, K.M., Zachara, J.M., and Fredrickson, J.K. (2012) Mtr extracellular electron-transfer pathways in Fe(III)-reducing or Fe(II)-oxidizing bacteria: a genomic perspective. Biochem Soc Trans 40: 1261-1267.
Straub, K.L., Benz, M., Schink, B., and Widdel, F. (1996) Anaerobic, nitrate-dependent microbial oxidation of ferrous iron. Appl Environ Microbiol 62: 1458-1460.
Publications
-
He, S., Tominski, C., Kappler, A., Behrens, S., & Roden, E. E. (2016). Metagenomic Analyses of the Autotrophic Fe(II)-Oxidizing, Nitrate-Reducing Enrichment Culture KS. Applied and Environmental Microbiology, 82(9), 2656–2668. doi:10.1128/aem.03493-15
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Eric Boyd
Co-Investigator
Shaomei He
Collaborator
-
RELATED OBJECTIVES:
Objective 2.1
Mars exploration.
Objective 3.2
Origins and evolution of functional biomolecules
Objective 4.1
Earth's early biosphere.
Objective 5.1
Environment-dependent, molecular evolution in microorganisms
Objective 5.3
Biochemical adaptation to extreme environments