2011 Annual Science Report
 Georgia Institute of Technology
Reporting | SEP 2010 – AUG 2011
Georgia Institute of Technology
Reporting | SEP 2010 – AUG 2011
Assessing the Role of Chance vs. Necessity in Evolution: Experimental Evolution of Ancient Proteins in E.coli
Project Summary
The goal of our current work is to create a “Bacterial Jurassic Park” in the laboratory. Our system combines synthetic biology (paleogenetics) with experimental evolution whereby we insert ancient genes into a modern bacterial genome. Replacing an essential bacterial gene with its ancient counterpart allows me to initiate a struggle for existence in these microbial populations since the ancient gene is maladapted to modern environments. Observing the real-time evolution of these resurrected genes as they adapt to the conditions of modern bacteria therefore allows me to monitor evolution in action (1-6).
Project Progress
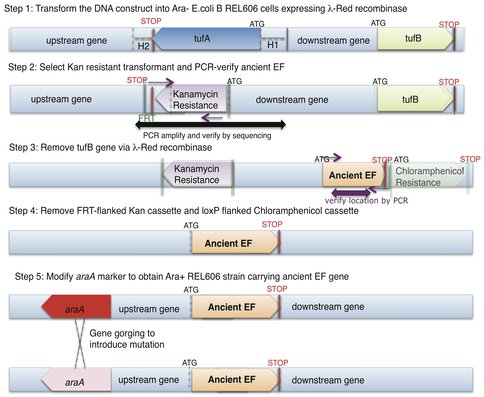
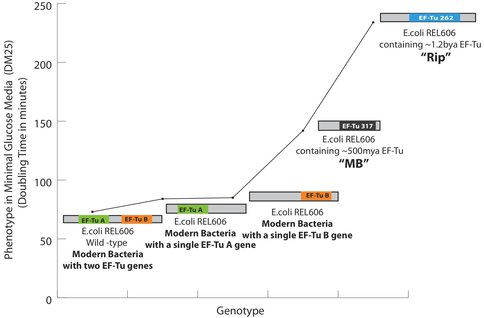
Ancestral EF-Tu proteins, resurrected previously by Eric Gaucher (7) exhibit a range of thermostability profiles (from 39.1 °C to 73.3 °C). During the first quarter of 2011, we finalized an experimental methodology that allows us to efficiently delete the two copies of EF-Tu genes present in modern E. coli and insert an ancestral EF-Tu gene in the precise chromosomal location of one of the modern genes. We have accomplished this goal and to date we have placed two ancient EF-Tus (the ~500 million years old, with the melting temperature of 39.1 °C and the 1.3 billion years old, with the melting temperature of 60.2 °C) (named “MB” and “Rip” respectively) for their modern counterpart within E.coli that grows at 37°C. (See Figure 1 for the final recombineering outline) Considering the highly adaptive properties of E. coli and EF-Tu, and the essentiality of EF-Tu for cell viability since this protein shuttles aminoacylated-tRNAs to the ribosome, our replacement of modern genes for ancient genes has indeed provided a scenario for rapid adaptive evolution to take within modern E. coli. (See Figure 2 for the effects of these replacements on the bacterial doubling time). Completion of this aim marks the first time ancient genes have been incorporated inside a bacterial genome and the bacteria containing these ancient genes exhibit a lower fitness than the strain hosting the modern form of the gene (Figure 2 & 3).
Once we obtained and verified the bacteria containing ancient gene 317 (MB strain), we prepared this particular construct for evolution, selection and competition. For this purpose, we have utilized a technique called ‘gene gorging’ (8) and engineered a neutral arabinose marker (Ara /-) gene in order to obtain pink (Ara) and red (Ara-) variants of the MB Strain. This phenotype allows us to determine the results of competition assays on tetrazolium-arabinose (TA) plates. We performed various experiments (obtaining growth curves, fitness effects) to validate the neutrality of this engineering on the phenotype of MB strain.
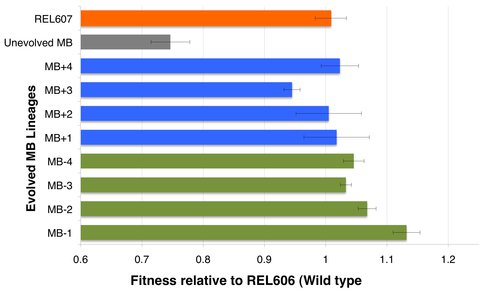
We have been evolving 4 flasks of Ara- and 4 flasks of Ara+ MB strains in 9.9 mL minimal glucose media (DM25) at 37 degrees for over 5 months now. Every day, after 24 hours of growth, we transfer 100 μL of each of the evolved culture into new fresh DM25 media. We calculate the fitness values of these evolved lineages every 100 generations (~16 days) by competition with REL606 (wild-type) as well as MB (unevolved) strains. We also measure the growth curves of the evolved lineages and calculate the effect on doubling times. Here, for simplicity, we are reporting the fitness values of MB strains at ‘generation 500’ (Figure 3). (Values obtained by competing 5-replicate population of each MB strain with REL606 (wild type) for 1 to 4 days, data obtained during Betul Arslan’s visit to Rich Lenski’s lab at MSU (supported by a NAI Research Scholarship). Competition data shows the adaptive evolution of MB lineages, which increases the fitness of these strains by 20% in less than 4 months of evolution. In order to understand the underlying genetic causes of fitness increase, we perform gene and genome sequencing to evolved strains. A preliminary sequencing data shows that one of the MB lineages experienced duplication in the promoter region of modern EF-Tu gene around 300 generation, suggesting that a fitness optimization through regulatory region modification. On going work includes understanding how much does this duplication in the promoter region actually increase the ancient EF-Tu expression and whether such optimization did/will affect the evolutionary trajectory taken by this lineage. While so far we have not identified any mutations in the ancient EF-Tu gene in any MB lineage, currently we are applying whole-genome sequencing to 8 MB-generation-500 strains to understand the effect of evolution in the context of the entire organism (at MSU sequencing facilities) (9-10). Such whole-genome sequencing will be performed every 500-generations. Other on-going work includes dissecting the phenotypic properties of evolved MB lineages (both in population and molecular level), as well as preparing the REL606 strain with a ~1.3 billion year old EF-Tu, Rip, for evolution through gene gorging as described above.
References
1. Barrick, J.E., et al., Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature, 2009. 461(7268): p. 1243-7.
2. Blount, Z.D., C.Z. Borland, and R.E. Lenski, Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Natl Acad Sci U S A, 2008. 105(23): p. 7899-906.
3. Cooper, T.F., D.E. Rozen, and R.E. Lenski, Parallel changes in gene expression after 20,000 generations of evolution in Escherichiacoli. Proc Natl Acad Sci U S A, 2003. 100(3): p. 1072-7.
4. Elena, S.F. and R.E. Lenski, Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet, 2003. 4(6): p. 457-69.
5. Khan, A.I., et al., Negative epistasis between beneficial mutations in an evolving bacterial population. Science, 2011. 332(6034): p. 1193-6.
6. Lenski, R.E., J.E. Barrick, and C. Ofria, Balancing robustness and evolvability. PLoS Biol, 2006. 4(12): p. e428.
7. Gaucher, E.A., S. Govindarajan, and O.K. Ganesh, Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature, 2008. 451(7179): p. 704-7.
8. Herring, C.D., J.D. Glasner, and F.R. Blattner, Gene replacement without selection: regulated suppression of amber mutations in Escherichia coli. Gene, 2003. 311: p. 153-63.
9. Chou, H.H., et al., Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science, 2011. 332(6034): p. 1190-2.
10. Nahum, J.R., B.N. Harding, and B. Kerr, Evolution of restraint in a structured rock-paper-scissors community. Proc Natl Acad Sci U S A, 2011. 108 Suppl 2: p. 10831-8.
Outline of the complete recombineering (recombinase mediated genetic engineering) and ‘gene gorging’ methodology obtained for this project. Two genes, tufA and tufB, (varying by just one amino acid) code for EF-Tu in REL606 E. coli. Precise replacement of endogenous EF-Tu requires both chromosomal tufA and tufB to be disrupted.
Phenotypic effects of the replacement of a modern EF-Tu gene with an ancient EF-Tu gene in bacteria: All measurements are performed at 37 °C in minimal glucose media. Two genes, shown as EF-Tu A and EF-Tu B encode for EF-Tu protein in bacteria. Deletion of the either copy increases the cell doubling time by ~30%. The increase in the doubling time after the replacement of EF-Tu B with EF-Tu 317 (‘MB’, 500 mya EF-Tu) and EF-Tu 262 (‘Rip’, 1.3Ga EF-Tu) in a strain that lacks EF-Tu A is shown. For the first time, ancient genes have been incorporated into a bacterial genome and further are shown to decrease the bacterial fitness as measured by an increase in the cell doubling time. Specifically, MB317 doubling time is 112 minutes, Rip262 doubling time is ~220 minutes, the modern bacterial cells with EF-Tu B gene double every other 85 minutes whereas the modern bacterial cells containing both of the EF-Tu A and B genes double every other 72 minutes in minimal glucose media at 37 degrees.
Figure 3 specifically shows the fitness values for MB strain (unevolved), 8 evolved MB lineages (generation 500) and the control REL607 relative to (in competition with) REL606 (wild type) strain.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Betül Kacar
Postdoc
-
RELATED OBJECTIVES:
Objective 4.2
Production of complex life.