2010 Annual Science Report
 Montana State University
Reporting | SEP 2009 – AUG 2010
Montana State University
Reporting | SEP 2009 – AUG 2010
Subglacial Methanogenesis and Implications for Planetary Carbon Cycling
Project Summary
Methanogens are thought to be among the earliest emerging life forms. Today, the distribution of methanogens is narrowly constrained, due in part to the energetics of the reactions which support this functional class of organism (namely carbon dioxide reduction with hydrogen and acetate fermentation). Methanogens utilize a number of metalloenzymes that have active site clusters comprised of a unique array of metals. The goals of this project are 1) identifying a suite of biomarkers indicative of biological CH4 production 2). quantifying the flux of CH~4~ from sub-ice systems and 3). developing an understanding how life thrives at the thermodynamic limits of life. This project represents a unique extension of the ABRC and bridges the research goals of several nodes, namely the JPL-Icy Worlds team and the ASU-Follow the Elements team.
Project Progress
Various genetic, biochemical, and geochemical biomarkers unique to methanogenic archaea including 16S rRNA and methyl Coenzyme M (CoM) reductase genes, glycerol dialkyl glycerol tetraether lipids, CoM, and methane (CH4) have been identified in association with the subglacial sediments beneath Robertson Glacier (RG), Canada, indicating the presence of an active assemblage of methanogens. To specifically examine the metabolisms that are supporting methanogenesis, sediment-containing mesocosms were designed that contain various carbon source amendments as well as specific metabolic inhibitors of methaogenic metabolisms. Mesocosm gas-phase CH4 and CoM quantification and qPCR of specific 16S rRNA genes are being monitored as a function of incubation time. These data will be used to estimate the productivity of substrate utilization in the phylogenetically-distinct methanogen lineages that inhabit the subglacial environment at RG.
A collaboration between the ASU-Follow the Elements team has been initiated to examine the geochemistry of the subglacial environment. This data will be used to predict the energetic feasibility of various metabolisms in the subglacial environment, including various forms of methanogenesis. These predictive data will be used to evaluate the empirical data derived from the mesocosm studies.
The CH4 production and rates determined from mesocosm studies has been used to estimate CH4 flux from sub-ice environments, both on Earth and on Mars. These values were then used to estimate the potential role of subglacial methanogenesis in driving climate change during the Laurentide glacial cycle, and to evaluate the plausibility of sub-ice methanogenesis in generating the CH4 plumes released from the Martian subsurface during summer 2003.
As part of this node of the ABRC project, NPP fellow Boyd has given numerous seminars (NAS Committee on the Origin and Evolution of Life, NAI-EC meeting, East Carolina University astrobiology seminar series, Montana State University’s Thermal Biology Institute seminar series). In addition, Boyd will be presented two talks at the upcoming American Geophysical Union conference and is organizing a session at the upcoming AbSciCon conference.
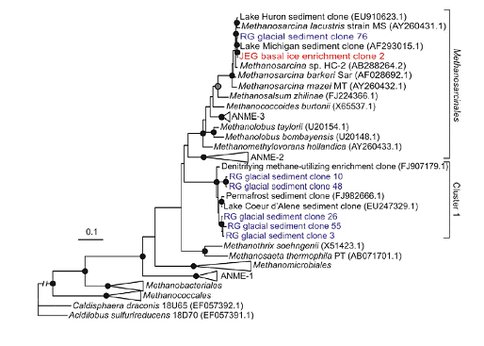
The phylogenetic position of archaeal 16S rRNA gene sequences recovered from RG (blue) and from an enrichment culture from JEG (red) and reference sequences were determined using PhyML. The tree was rooted with 16S rRNA genes from Caldisphaera draonis 18U65 and Acidilobus sulfurireducens 18D70, both of which are members of the Crenarchaeota. Black circles at nodes denote >90% bootstrap support (BS), grey circles at nodes denote >80% BS, open circles denote >70% BS and no symbol denotes <70% BS. Bar equals two substitutions per 10 sequence positions. The branch length of the out group was cropped to conserve space.
Robertson Glacier (RG), Kananaskis Country, Alberta, Canada looking south (July 2008). Inset, east stream sampling site in September 2009 (photo credit: Everett Shock). Arrows indicate sub-ice sampling locations
Publications
-
Boyd, E. S., Lange, R. K., Mitchell, A. C., Havig, J. R., Hamilton, T. L., Lafreniere, M. J., … Skidmore, M. (2011). Diversity, Abundance, and Potential Activity of Nitrifying and Nitrate-Reducing Microbial Assemblages in a Subglacial Ecosystem. Applied and Environmental Microbiology, 77(14), 4778–4787. doi:10.1128/aem.00376-11
-
Boyd, E. S., Skidmore, M., Mitchell, A. C., Bakermans, C., & Peters, J. W. (2010). Methanogenesis in subglacial sediments. Environmental Microbiology Reports, 2(5), 685–692. doi:10.1111/j.1758-2229.2010.00162.x
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Mark Skidmore
Project Investigator
John Peters
Co-Investigator
Eric Boyd
Postdoc
Matthew Urschel
Doctoral Student
Rachel Lange
Undergraduate Student
-
RELATED OBJECTIVES:
Objective 2.1
Mars exploration.
Objective 5.1
Environment-dependent, molecular evolution in microorganisms
Objective 5.2
Co-evolution of microbial communities
Objective 5.3
Biochemical adaptation to extreme environments
Objective 7.1
Biosignatures to be sought in Solar System materials
Objective 7.2
Biosignatures to be sought in nearby planetary systems