2010 Annual Science Report
 Montana State University
Reporting | SEP 2009 – AUG 2010
Montana State University
Reporting | SEP 2009 – AUG 2010
Structure, Reactivity, and Biosynthesis of Cataylic Iron-Sulfur Clusters
Project Summary
We are examining the biosynthesis of complex iron-sulfur cluster to determine the specific chemistry associated with modifying iron-sulfur motifs in biology for different functions. We then relate the chemistry associated with these modifying reactions to reactions that could potentially modify iron-sulfur mineral motifs in the early non-living Earth to promote analogous reactivity.
Project Progress
During the current funding period we have made tremendous progress in the area of [FeFe]-hydrogenase H cluster biosynthesis and nitrogenase biosynthesis. These results have really changed the way in which we think about iron-sulfur enzyme evolution and have laid the groundwork for the development of a completely new experimental line described in the report (see Molecular Paleontology of iron-sulfur enzymes) probing evolution directly with parallel phylogenetic studies on structural and biosynthetic gene products in tandem. Another new line developed from the results this funding period is a thrust area devoted to the evolution and diversity of racial SAM enzyme reactivity also now described as an independent project in the report. Some of the key work reported already this year in the literature or related to the mechanism of H cluster assembly on a scaffold, the role radical SAM enzyme in the synthesis of unique nonprotein ligands and the mechanism of cluster assembly and stepwise maturation on the [FeFe]-hydrogenase structural proteins. We have also examined in collaborative work an reactive intermediate in nitrogen iron-molybdenum cofactor biosynthesis produced by the modification of an iron-sulfur cluster by a radical SAM enzyme and the introduction of a unique non protein ligand. Together these results reveal strong unifying themes in the biosynthesis and maturation of the complex iron-sulfur cluster active sites of Mo-nitrogenases and [FeFe]-hydrogenases. In work published in the Proceedings of the National Academy of Sciences we reported that as in the case of the nitrogenase iron-molydenum cofactor, the [FeFe]-hydrogenase H cluster requires a protein scaffold for complex cluster assembly. In work published in the Journal of the American Chemical Society and in Angewandte Chemie we have been able to show that the unique carbon monoxide and cyanide ligands to Fe in [FeFe]-hydrogenase result from the radical based cleavage of the amino acid tyrosine. This served to prove our previous controversial proposal that carbon monoxide and cyanide ligands could be produced in a concerted manner from amino acid radical decomposition. The results are important and represent biology own version of “ligand assisted catalysis” which has been described to occur when small organics potentially produced by mineral reactivity feed back and directly modify mineral to support new reactivity. Additionally in work published in Nature, through the structural characterization of a key intermediate in cluster biosynthesis we have been able to define the stepwise pathway of cluster biosynthesis.
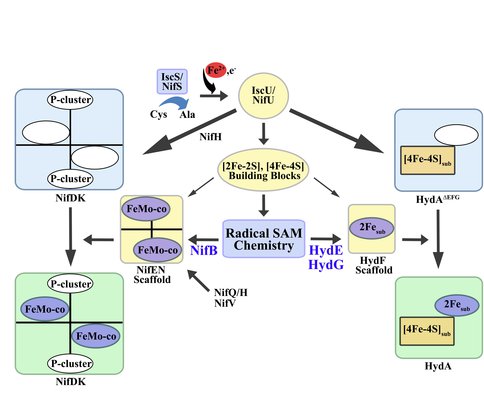
Cysteine desulfurases and scaffold proteins responsible for generalized iron-sulfur cluster assembly (IscS and IscU) and those common to nitrogen fixing organisms (NifS and NifU) synthesize the iron-sulfur cluster building blocks of the H-cluster of [FeFe] hydrogenase (right) and the P-cluster of nitrogenase (left). Further, IscS/NifS and IscU/NifU supply standard iron-sulfur clusters that feed into the biosynthetic pathway resulting in complex cofactor assembly. Specifically, NifS and NifU provide NifB with its iron sulfur core that becomes modified to NifB-co, whereas IscS and IscU presumably supply HydF with a [2Fe-2S] core that becomes modified to the 2Fe subcluster of the H-cluster. The modifications of these standard iron-sulfur clusters are accomplished through the action of radical SAM chemistry coupled to the presence of specific scaffold proteins in both nitrogenase and hydrogenase systems.
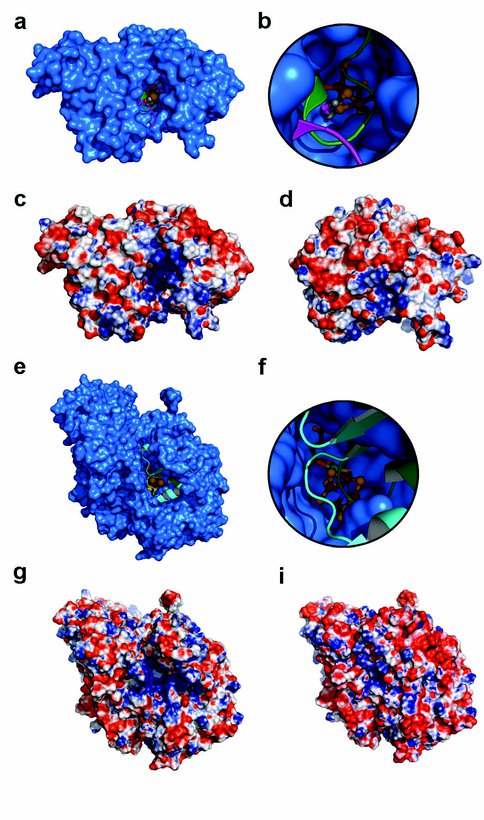
Surface representation of H cluster 2Fe subcluster deficient hydrogenase intermediate in assembly (blue) with superimposed native hydrogenase shown in ribbon represention. The two loop regions in CpI with major structural rearrangement (green and purple) overlay the channel present in H cluster biosynthetic intermediate prior to cluster insertion. b, Zoom down the channel of the intermediate to active site area where the 2Fe subcluster from superimposed CpI resides at the end (ball and stick represention). c, Electrostatic surface representation of the 2Fe subcluster deficient hydrogenase calculated with the PyMol plugin APBS. Negative charge (-10 kT) is red and positive charge (10 kT) is blue. d, Electrostatic surface representation on the catalytic domain of native hydrogenase in the same orientation as the 2Fe subcluster deficient hydrogenase intermediate and calculated with identical parameters as c. e, Surface representation of FeMo-co deficient MoFe protein (blue) with superimposed active MoFe protein from shown in ribbon representation. Regions showing major structural rearrangement in the MoFe protein are colored cyan and green and overlay the channel leading to the FeMo-co site. f, Zoom down the channel to the active site area where the FeMo-co from superimposed active MoFe protein resides (ball and stick representation). g, Electrostatic surface representation of FeMo-co deficient MoFe protein. i, Electrostatic surface representation of active MoFe protein in the same orientation. Both surface representations were calculated with identical parameters as c.
Publications
-
Driesener, R. C., Challand, M. R., McGlynn, S. E., Shepard, E. M., Boyd, E. S., Broderick, J. B., … Roach, P. L. (2010). -Hydrogenase Cyanide Ligands Derived From S-Adenosylmethionine-Dependent Cleavage of Tyrosine. Angewandte Chemie International Edition, 49(9), 1687–1690. doi:10.1002/anie.200907047
-
McGlynn, S. E., Boyd, E. S., Shepard, E. M., Lange, R. K., Gerlach, R., Broderick, J. B., & Peters, J. W. (2009). Identification and Characterization of a Novel Member of the Radical AdoMet Enzyme Superfamily and Implications for the Biosynthesis of the Hmd Hydrogenase Active Site Cofactor. Journal of Bacteriology, 192(2), 595–598. doi:10.1128/jb.01125-09
-
McGlynn, S. E., Mulder, D. W., Shepard, E. M., Broderick, J. B., & Peters, J. W. (2009). Hydrogenase cluster biosynthesis: organometallic chemistry nature’s way. Dalton Trans., None(22), 4274. doi:10.1039/b821432h
-
Mulder, D. W., Boyd, E. S., Sarma, R., Lange, R. K., Endrizzi, J. A., Broderick, J. B., & Peters, J. W. (2010). Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydAΔEFG. Nature, 465(7295), 248–251. doi:10.1038/nature08993
-
Mulder, D. W., Ortillo, D. O., Gardenghi, D. J., Naumov, A. V., Ruebush, S. S., Szilagyi, R. K., … Peters, J. W. (2009). Activation of HydA ΔEFG Requires a Preformed [4Fe-4S] Cluster. Biochemistry, 48(26), 6240–6248. doi:10.1021/bi9000563
-
Peters, J. W., Boyd, E. S., D’Adamo, S., Mulder, D. W., Therien, J., & Posewitz, M. C. (2012). Hydrogenases, Nitrogenases, Anoxia, and H2 Production in Water-Oxidizing Phototrophs. Algae for Biofuels and Energy, None, 37–75. doi:10.1007/978-94-007-5479-9_3
-
Sarma, R., Barney, B. M., Keable, S., Dean, D. R., Seefeldt, L. C., & Peters, J. W. (2010). Insights into substrate binding at FeMo-cofactor in nitrogenase from the structure of an α-70Ile MoFe protein variant. Journal of Inorganic Biochemistry, 104(4), 385–389. doi:10.1016/j.jinorgbio.2009.11.009
-
Shepard, E. M., Boyd, E. S., Broderick, J. B., & Peters, J. W. (2011). Biosynthesis of complex iron–sulfur enzymes. Current Opinion in Chemical Biology, 15(2), 319–327. doi:10.1016/j.cbpa.2011.02.012
-
Shepard, E. M., Duffus, B. R., George, S. J., McGlynn, S. E., Challand, M. R., Swanson, K. D., … Broderick, J. B. (2010). -Hydrogenase Maturation: HydG-Catalyzed Synthesis of Carbon Monoxide. Journal of the American Chemical Society, 132(27), 9247–9249. doi:10.1021/ja1012273
-
Shepard, E. M., McGlynn, S. E., Bueling, A. L., Grady-Smith, C. S., George, S. J., Winslow, M. A., … Broderick, J. B. (2010). Synthesis of the 2Fe subcluster of the [FeFe]-hydrogenase H cluster on the HydF scaffold. Proceedings of the National Academy of Sciences, 107(23), 10448–10453. doi:10.1073/pnas.1001937107
-
Soboh, B., Boyd, E. S., Zhao, D., Peters, J. W., & Rubio, L. M. (2010). Substrate specificity and evolutionary implications of a NifDK enzyme carrying NifB-co at its active site. FEBS Letters, 584(8), 1487–1492. doi:10.1016/j.febslet.2010.02.064
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Joan Broderick
Project Investigator
John Peters
Project Investigator
Eric Shepard
Postdoc
Trevor Beard
Doctoral Student
Nicholas Boswell
Doctoral Student
Ben Duffus
Doctoral Student
Shawn McGlynn
Doctoral Student
David Mulder
Doctoral Student
Kevin Swanson
Doctoral Student
-
RELATED OBJECTIVES:
Objective 3.1
Sources of prebiotic materials and catalysts
Objective 3.2
Origins and evolution of functional biomolecules
Objective 3.3
Origins of energy transduction
Objective 3.4
Origins of cellularity and protobiological systems
Objective 4.1
Earth's early biosphere.
Objective 7.1
Biosignatures to be sought in Solar System materials
Objective 7.2
Biosignatures to be sought in nearby planetary systems