2010 Annual Science Report
 NASA Ames Research Center
Reporting | SEP 2009 – AUG 2010
NASA Ames Research Center
Reporting | SEP 2009 – AUG 2010
Origins of Functional Proteins and the Early Evolution of Metabolism
Project Summary
The main goal of this project is to identify critical requirements for the emergence of biological complexity in early habitable environments by examining key steps in the origins and early evolution of functional proteins and metabolic reaction networks. In particular, we investigate whether protein functionality can arise from an inventory of polymers with random sequences that might have naturally existed in habitable environments. We attempt the first demonstration of multiple origins of a single enzymatic function, and investigate experimentally how primordial proteins could evolve through the diversification of their structure and function. Building on this work and on our knowledge of ubiquitous protocellular functions and constraints of prebiotic chemistry, we conduct computer simulations aimed at elucidating fundamental principles that govern coupled evolution of early metabolic reactions, their catalysts and transport across cell walls.
Project Progress
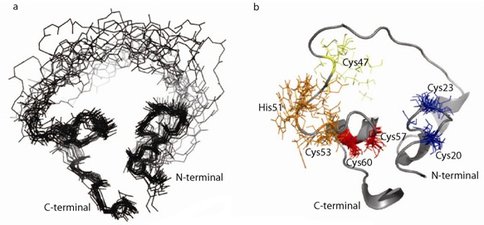
We have been characterizing an artificial enzyme that joins two RNA fragments. This enzyme was previously generated de novo from a large library of proteins based on a zinc finger protein scaffold with randomized loops. Structural investigations by Nuclear Magnetic Resonance (NMR) have generated sufficient data to model an ensemble of its structures (Fig. 1a). The protein has a highly compact core composed of two regions connected by a flexible loop and very flexible ends. NMR data suggest that two zinc-binding sites are located in the compact core. The protein structure is unrelated to the original scaffold and has no homology to other known structures. This indicates that (1) protein evolution through a modest number of mutations can yield both novel functions and novel structures, and (2) structures and functions not found in contemporary organisms are possible. We have generated over 20 mutant proteins (Fig. 1b) to identify the zinc-binding sites and the amino acids involved in catalysis.
Figure 1.. a) Ensemble of 20 lowest energy structures of the ligase protein based on NMR data. b) Mutations mapped onto the structure. Insoluble mutants are shown in red (C57S and C60S), mutants causing structural alterations in blue (C20 and C23), loss of function mutants preserving the original structure is in orange (H51 and C53) and a mutant that both retains the original CD spectrum and partial ligase activity is shown in yellow (C47S or C47R).
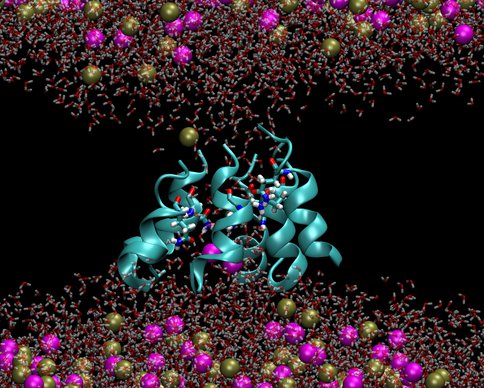
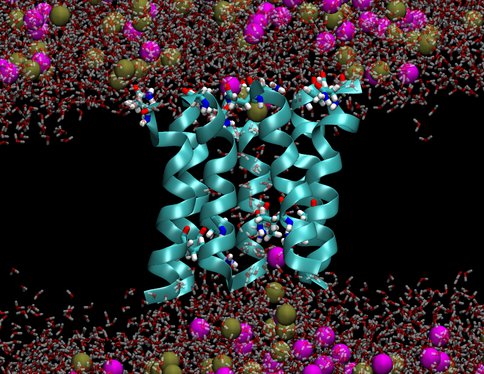
We investigated two models of protobiological protein ion channels, antiamoebin and trichotoxin. They facilitate ion transport across cell walls needed for establishing osmotic equilibria, energy transduction and stability of macromolecules in protocells. Peptides forming the channels consist of only 14-16 residues and contain non-standard amino acids that were common on the early earth. By comparing calculated and measured conductance we determined the channel structures. Despite their simplicity the channels are quite efficient in increasing rates of ion transport through membranes, a property that is essential for preventing the buildup of osmotic stresses. The specific identity of many amino acids in these channels is not important, as long as their polarity is conserved, a convenient property at the origins of life. Our results support a view that the first ion channels were similar to the channels studied here.
Figure 2a..
Figure 2b.. Pictures from molecular dynamics simulations of transmembrane ion channels formed by (1) a hexameric bundle of antiamoebin and (2) a heptameric bundle of trichotoxin. The protein backbones are shown as light blue ribbons, the glutamine residues are shown as bonds with C, O, N and H atoms colored light blue, red, dark blue and white, respectively, water molecules are shown as transparent red and white bonds, and the K+ and Cl- ions are shown as spheres colored gold and magenta, respectively. The phospholipids forming the membrane have been removed for clarity.
Publications
-
Golynskiy, M. V., & Seelig, B. (2010). De novo enzymes: from computational design to mRNA display. Trends in Biotechnology, 28(7), 340–345. doi:10.1016/j.tibtech.2010.04.003
-
Pohorille, A., Jarzynski, C., & Chipot, C. (2010). Good Practices in Free-Energy Calculations. J. Phys. Chem. B, 114(32), 10235–10253. doi:10.1021/jp102971x
-
Wei, C., & Pohorille, A. (2011). Permeation of Nucleosides through Lipid Bilayers. J. Phys. Chem. B, 115(13), 3681–3688. doi:10.1021/jp112104r
-
Wilson, M. A., Wei, C., Bjelkmar, P., Wallace, B. A., & Pohorille, A. (2011). Molecular Dynamics Simulation of the Antiamoebin Ion Channel: Linking Structure and Conductance. Biophysical Journal, 100(10), 2394–2402. doi:10.1016/j.bpj.2011.03.054
- Pohorille, A. (2010). Emerging properties in the origins of life and Darwinian evolution. Orig. Life Evol. Biosphere, 40: 384-386.
- Pohorille, A. (2010). Was the Emergence of Life on Earth a Likely Outcome of Chemical Evolution? Orig. Life Evol. Biosphere, 40: 362-365.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Burckhard Seelig
Co-Investigator
Michael Wilson
Co-Investigator
James Lake
Collaborator
Milan Mijajlovic
Postdoc
Chenyu Wei
Research Staff
Aleardo Morelli
Graduate Student
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules
Objective 3.4
Origins of cellularity and protobiological systems