2009 Annual Science Report
 Georgia Institute of Technology
Reporting | JUL 2008 – AUG 2009
Georgia Institute of Technology
Reporting | JUL 2008 – AUG 2009
Ribosome Paleontology
Project Summary
We are establishing method to determine chronologies of ancient ribosomal evolution. One method, just published, uses structure-based and sequence-based comparisons of the LSUs of Haloarcula marismortui and Thermus thermophilus, along with an “onion approximation”. The results suggest that the conformation and interactions of both RNA and protein change, in an observable manner, over evolutionary time.
Project Progress
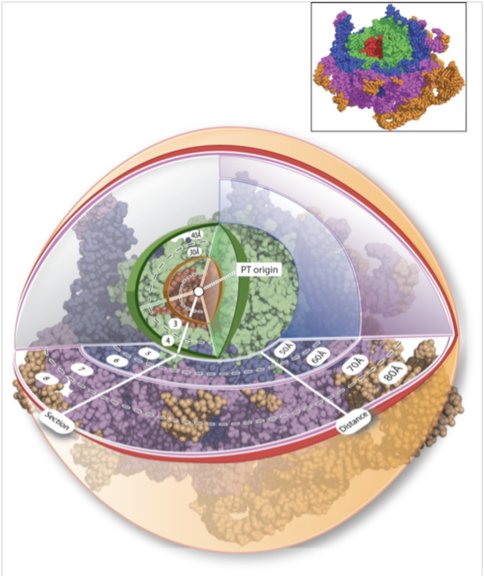
We have described a method to establish chronologies of ancient ribosomal evolution. This method combines the onion approximation^1^ with our previous methods2-7 of structural analysis of large RNA complexes. We have combined structure-based and sequence-based comparison of the large subunits (LSUs) of Haloarcula marismortui and Thermus thermophilus. These are the highest resolution ribosome structures available and represent disparate regions of the evolutionary tree. We have sectioned the superimposed LSUs into concentric shells, like an onion (Figure 1)
Figure 1. Figure 1. Ribosomal paleontology. The Haloarcula marismortui (and Thermus thermophilus, not shown) LSUs have been sectioned into concentric shells, with the origin at the site of peptidyl transfer (the PT-origin). The radius of any shell is 10 Å greater than the radius of the preceding shell. 23S rRNAHM is red in shells 1 and 2, green in shells 3 and 4, blue in shells 5 and 6, and purple in shells 7 and 8. Additional shells are shown in orange. Atoms are represented in spacefill. Ribosomal proteins, ions, and water molecules are omitted for clarity.
Using the site of peptidyl transfer as the origin (the PT-origin). This spherical approximation, combined with a shell-by-shell comparison, captures significant information about ribosomal evolution, suggesting that the conformation and interactions of both RNA and protein are changing over evolutionary time. The tendency of macromolecules (protein and RNA) to assume regular secondary structural elements such as A-form helices with Watson–Crick base pairs (RNA) and α-helices and β-sheets (protein) is low at early time points but increases as time progresses. The non-secondary structures of ribosomal protein segments near the PT-origin suggest that they may be molecular fossils of the peptide ancestors of ribosomal proteins. The rRNA in the inner shells suggests that initial formation of the PTC may have involved Mg2+-mediated assembly of at least partially single-stranded RNA oligomers or polymers. As one moves from center to periphery, proteins appear to replace magnesium ions.
Publications
-
Hsiao, C., & Williams, L. D. (2009). A recurrent magnesium-binding motif provides a framework for the ribosomal peptidyl transferase center. Nucleic Acids Research, 37(10), 3134–3142. doi:10.1093/nar/gkp119
-
Hsiao, C., Mohan, S., Kalahar, B. K., & Williams, L. D. (2009). Peeling the Onion: Ribosomes Are Ancient Molecular Fossils. Molecular Biology and Evolution, 26(11), 2415–2425. doi:10.1093/molbev/msp163
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Chiaolong Hsiao
Postdoc
Catherine Trippe
Undergraduate Student
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules